Fig. 4.
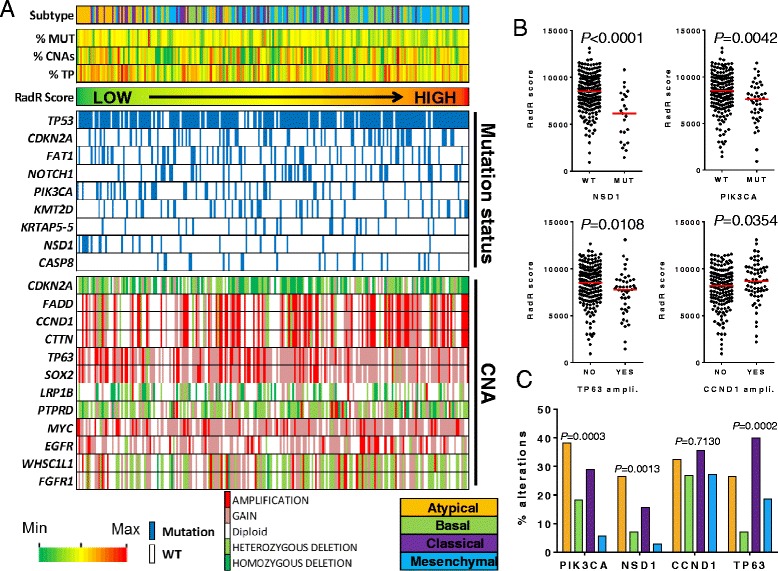
Integrative analysis of the radioresistance score with known recurrent genomic alterations in 220 human papillomavirus (HPV)-negative head and neck squamous cell carcinoma (HNSCC) from The Cancer Genome Atlas (TCGA). a Patients are represented by columns while specific variables are represented by rows. Tumors are ranked by the radioresistance (RadR) score computed in each tumor sample. The molecular class, the percentage of copy number alterations (%CNAs), the mutation count (%MUT), and the tumor purity (%TP) are shown for each patient. We selected known recurrently altered genes at the genomic level according to the previous publication by the TCGA network [26]. CNAs from GISTIC as well as somatic mutations of recurrently altered genes with a frequency > 10% are shown. A color range from green to red was used to represent increasing numerical values. b Using a Mann–Whitney test, the RadR score was compared between samples with NSD1 and PIK3CA mutations compared to wild type samples, as well as between samples which harbored CCND1 and TP63 amplifications and those which did not. c Percentage of genomic alterations of NSD1, PIK3CA (mutations), and CCND1 and TP63 (amplifications) were compared across molecular subtypes of HPV-negative of HNSCC
