Abstract
Background
Molecular biomarkers capable of predicting recurrence patterns and prognosis are helpful for risk stratification and providing appropriate treatment to patients with hepatocellular carcinoma (HCC). In this study, we focused on G protein-coupled receptor 155 (GPR155), a cell surface signaling protein, as a candidate biomarker.
Methods
We analyzed GPR155 expression, DNA methylation, and copy number in HCC cell lines. The clinical significance of GPR155 expression was evaluated using 144 pairs of surgically resected liver and normal tissues with subgroup analysis based on hepatitis virus infection.
Results
GPR155 mRNA expression levels were differential and were decreased in 89% of HCC cell lines. No DNA methylation was detected, whereas copy number alterations were present in five (56%) HCC cell lines. GPR155 mRNA expression level was independent of background liver status and significantly lower in HCC tissues than corresponding normal liver tissues. The expression patterns of GPR155 protein by immunohistochemical staining were significantly associated with those of GPR155 mRNA. Downregulation of GPR155 was significantly associated with more aggressive HCC phenotypes including high preoperative α-fetoprotein, poor differentiation, serosal infiltration, vascular invasion, and advanced disease stage. Patients with downregulation of GPR155 were more likely to have worse prognosis after curative resection irrespective of hepatitis virus infection. Patients who experienced extrahepatic (distant) recurrences had significantly lower GPR155 expression than those with intrahepatic (liver confined) recurrences.
Conclusions
Downregulation of GPR155 may serve as a prognosticator that also predicts initial recurrence sites independent of hepatitis virus infection.
Electronic supplementary material
The online version of this article (10.1186/s12885-017-3629-2) contains supplementary material, which is available to authorized users.
Keywords: Hepatocellular carcinoma, GPR155, Expression, Recurrence, Biomarker
Background
Hepatocellular carcinoma (HCC) ranks at the third most common cause of cancer-related death in the world [1, 2]. Although liver resection has been the mainstay of treatment for HCC, the recurrence rate after curative resection remains high at approximately 70% [2–4]. Complete cure of this disease is quite challenging even though various therapeutic modalities have been developed. A realistic initial goal is the establishment of methods for accurate risk stratification and prediction of recurrence sites after liver resection to provide appropriate perioperative management according to each individual patient’s circumstances [5]. The TNM classification system has been broadly employed as a tumor staging method to predict postoperative outcomes concisely but can be inaccurate [6, 7]. For example, patients with an earlier tumor stage sometimes have unfavorable prognosis. Extrahepatic recurrences, such as lung, bone, and brain metastases, can be a cause of an unexpected and rapidly deteriorating patient course; however, no methods for predicting the likelihood of extrahepatic recurrences of HCC are currently available [8, 9]. Conversely, some patients are long-term survivors after resection of advanced HCC without adjuvant therapy. To address these clinical issues, development of a novel molecular marker able to reflect potential characteristics of the tumor is required [10].
G protein-coupled receptors (GPCRs) are reportedly cell surface signaling proteins that have important roles in various physiological functions, and in initiation and progression of cancer [11]. The G protein-coupled receptor 155 gene (GPR155), present on 2q31.1, encodes a 97 kDa transmembrane receptor protein that is a member of the GPCR family [12]. Although there has been a report that GPR155 expression is suppressed in neoplasms of the thyroid, the oncologic roles of GPR155 in HCC remain unclear [13, 14]. We focus on GPR155 because it is recognized as a transmembrane marker possibly associated with the transport of growth factors and anticancer drugs, and no published data of GPR155 expression in HCC.
The aims of this study were to evaluate the clinical significance of GPR155 expression, explore the factors that regulate GPR155 transcription, and assess the performance of GPR155 as a potential prognosticator of HCC.
Methods
Sample collection
Human HCC cell lines Hep3B, HepG2, PLC/PRF/5, and SK-Hep1, and the control nontumorigenic epithelial cell line FHs74 were obtained from the American Type Culture Collection (Manassas, VA). HLE, HLF, HuH1, and HuH7 cells were obtained from the Japanese Collection of Research Bioresources Cell Bank (Osaka, Japan). HuH2 was from Aichi Cancer Center (Nagoya, Japan). Primary HCC tissues and corresponding non-cancerous tissues were collected from 144 patients who underwent liver resection at Nagoya University Hospital between January 1998 and January 2012. All tissue samples were frozen immediately after resection and diagnosed histologically as HCC. Postoperative follow-up included physical examinations, measurement of serum tumor markers every 3 months, and enhanced computed tomography every 6 months [15]. Treatment after recurrence included surgery, radiofrequency ablation, transcatheter arterial chemoembolization, and chemotherapy, according to tumor status and liver function.
Quantitative real-time RT-PCR (qRT-PCR)
Quantitative real-time reverse-transcription polymerase chain reaction (qRT-PCR) was used to determine the expression level of GPR155 mRNA. Primer sequences are shown in Supplementary Table 1. Total RNA (10 μg per sample) was isolated from nine HCC cell lines, FHs74 cells, and 144 pairs of clinical samples and a quality check for all RNA samples was conducted before generating complementary DNAs (cDNAs). The optical density was measured and the ratio of the absorbance at 260 and 280 nm ranged from 1.8 to 2.0 in all samples. cDNA was generated from 1 μg of total RNA using M-MLV Reverse Transcriptase (Thermo Fisher Scientific, Waltham, MA, USA) with 1 h incubation at 37 °C. qRT-PCR was performed using the SYBR Green PCR Core Reagents Kit (Applied Biosystems, Foster City, CA, USA) as follows: one cycle at 95 °C for 10 min, 40 cycles at 95 °C for 5 s, and 60 °C for 60 s, and included no-template samples as a negative control. Real-time detection of SYBR Green fluorescence was conducted using an ABI StepOnePlus Real-Time PCR System (Applied Biosystems). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA (TaqMan, GAPDH control reagents, Applied Biosystems) was quantified as an endogenous control in each sample for normalization [16]. The qRT-PCR reactions in each sample were performed in triplicate. The relative copy number of the mRNA was calculated in reference to standard curves (cloned 101 ~ 107 amplicons) established by our laboratory. The expression level of each sample is presented as the value of the GPR155 amplicon divided by that of GAPDH (Additional file 1: Table S1) [17].
Bisulfite sequence analysis
We conducted methylation analysis assuming the existence of DNA hypermethylation because GPR155 harbors a CpG island in its promoter region. Genomic DNA of the cell lines was treated with bisulfite for bisulfite sequence analysis [18]. After PCR amplification using specific primers shown in Additional file 1: Table S1, the PCR products were purified using a MiniElute PCR Purification Kit (Qiagen, Hilden, Germany) and 5 ng of PCR products mixed with 9.6 pmol of sense primer were sent to Eurofins Genomics Co. Ltd. (Tokyo, Japan) for sequencing.
Copy number analysis
Using purified genomic DNA obtained from HCC cell lines, DNA copy numbers were determined by the TaqMan Copy Number Assays (Applied Biosystems) to explore regulatory mechanisms of GPR155 expression other than DNA methylation. A total of 20 ng of genomic DNA was amplified with specific primer pairs according to the manufacturer’s protocol using an ABI StepOnePlus Real-Time PCR System (Applied Biosystems). Three assays were employed: upstream (assay ID: Hs01092594_cn, location: Chromosome 2, 175,351,658 in the exon 1 of GPR155 gene), midstream (assay ID: Hs01971174_cn, location: Chromosome 2, 175,335,170 in the exon 6 of GPR155 gene), and downstream (assay ID: Mn00059996_cn, location: Chromosome 2, 73,351,855 at overlaps intron 14 and exon 14 of GPR155 gene). Data were analyzed using CopyCallerTM Software (Life Technologies, Carlsbad, CA, USA) [19].
Immunohistochemical staining of GPR155 protein
Immunohistochemical staining was performed to determine the difference in GPR155 protein expression between HCC tissue and non-cancerous tissues in 60 specimens. Sections were incubated for 16 h at 4 °C with a rabbit polyclonal antibody raised against GPR155 (sc-137,511, Santa Cruz Biotechnology Inc., Dallas, TX, USA) diluted 1:200 in Antibody Diluent (Dako, Carpinteria, CA, USA). Sections were washed with phosphate buffered saline, followed by a 10-min incubation with biotinylated secondary antibody (SignalStain® Boost IHC Detection Reagent labelled by HRP, Cell Signaling Technology, Beverly, MA, USA). Antigen-antibody complexes were visualized by exposure of liquid 3, 3′-diaminobenzidine (Nichirei, Tokyo, Japan) for five minutes. Two independent observers evaluated the specimens in a blinded manner as follows: HCC > non-cancerous component, equivalent, or HCC < non-cancerous component [20].
Statistical analysis
Differences between data of two groups were evaluated using the Mann–Whitney test. The χ2 test was used to analyze the significance of the association between the expression levels of GPR155 mRNA and patients’ clinicopathologic parameters. Survival rates were calculated using the Kaplan–Meier method and differences in survival curves were evaluated using the log-rank test. All statistical analyses were performed using JMP 10 software (SAS Institute Inc., Cary, NC). A p value <0.05 was considered statistically significant.
Results
Expression, methylation, and copy number alteration of GPR155 in cell lines
GPR155 showed differential mRNA expression with decreased levels of expression in all HCC cell lines except for HuH1 compared with the control non-tumorigenic cell line FHs74 (Fig. 1a). No significant difference in expression levels of GPR155 mRNA was observed between differentiated and undifferentiated types. Bisulfite sequence analysis revealed no DNA methylation at the region amplified by our primers within the promoter of GPR155 gene (Fig. 1b). However, copy number alterations were detected in HuH2, HuH7, PLC/PRF/5, HuH1, and SK-Hep1 cells (Fig. 1a).
Fig. 1.
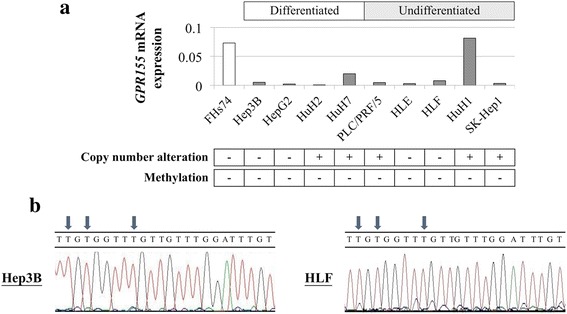
Analysis of expression, methylation, and copy number of GPR155 in cell lines. a GPR155 mRNA expression levels in HCC cell lines. Copy number alterations and methylation status of the GPR155 promoter are summarized in lower boxes. b Representative results of bisulfite sequence analysis. All CpG sites were converted to TG
Patient characteristics
The age of the 144 patients ranged from 34 to 84 years (median 65.5 years) and the male:female ratio was 121:23. Thirty-seven patients were infected with hepatitis B virus (HBV) and 80 patients with hepatitis C virus (HCV). The number of patients with normal liver, chronic hepatitis, and cirrhosis was 10, 82, and 52, respectively. Ninety, 37, and 17 patients were in stage I, II, or III, respectively, according to the Union for International Cancer Control (UICC) classification.
Analysis of GPR155 mRNA and protein expression in HCC tissues
GPR155 mRNA expression levels in non-cancerous tissues were comparable among patients with normal liver, chronic hepatitis, and cirrhosis (Fig. 2a). HCC tissues had significantly lower expression levels of GPR155 mRNA than the corresponding normal liver tissues (Fig. 2b). An inverse correlation between GPR155 expression levels in HCC tissues and preoperative serum α-fetoprotein was observed (Fig. 2c). The expression patterns of GPR155 protein were evaluated using immunohistochemical staining and two representative patients with reduced expression of GPR155 protein in the cytoplasm of cancer cells compared with non-cancerous cells are shown in Fig. 2d. The pattern of staining intensity of GPR155 protein between HCC and normal components was significantly associated with the qRT-PCR data (p < 0.001, Fig. 2d).
Fig. 2.
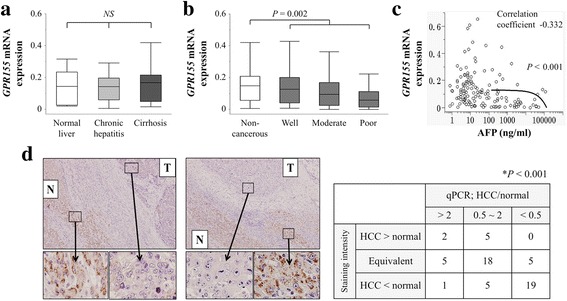
Analysis of GPR155 expression in clinical specimens. a There were no significant differences in GPR155 mRNA levels among non-cancerous tissues categorized by background uninvolved liver status. b GPR155 mRNA was expressed at lower levels in HCC tissues compared with corresponding non-cancerous tissues. c Correlation of GPR155 mRNA expression levels in HCC tissues with preoperative serum α-fetoprotein levels. d Detection of GPR155 protein in two representative patients. In both cases, cancerous tissues exhibited reduced expression compared with adjacent non-cancerous tissues (100× and 400× magnification). N, non-cancerous component; T, tumor component. A significant correlation between staining intensity and transcription patterns of GPR155 was observed
Clinical implications of GPR155 expression levels
Patients were categorized into two groups according to GPR155 expression level. Downregulation of GPR155 was defined as GPR155 expression level in HCC tissue ≤50% of that in the corresponding non-cancerous tissue. Downregulation of GPR155 was significantly associated with female sex, Pugh-Child’s classification B, α-fetoprotein >20 ng/mL, protein induced by vitamin K antagonists II >40 mAU/mL, poor differentiation, serosal infiltration, formation of capsule, infiltration to capsule, septum formation, vascular invasion, and advanced UICC stage (Table 1). The overall survival of patients with downregulation of GPR155 was significantly shorter than that of patients without downregulation of GPR155 (5-year survival rates 52% versus 72%, respectively, Fig. 3a). Disease-free survival was also shorter in patients with downregulation of GPR155 than in those without (2-year disease-free survival rates 41% versus 59%, respectively, Fig. 3b). Multivariable analyses were performed for both overall and disease-free survival and downregulation of GPR155 was not identified as an independent prognostic factor (Additional file 2: Table S2 and Additional file 3: Table S3).
Table 1.
Association between expression level of GPR155 mRNA and clinicopathological parameters in 144 patients with hepatocellular carcinoma
| Clinicopathological parameters | Downregulation of GPR155
(n = 57) |
Others (n = 87) |
p value |
|---|---|---|---|
| Age | 0.349 | ||
| < 65 year | 23 | 42 | |
| ≥ 65 year | 34 | 45 | |
| Gender | 0.013* | ||
| Female | 4 | 19 | |
| Male | 53 | 68 | |
| Background liver | 0.188 | ||
| Normal liver | 2 | 8 | |
| Chronic hepatitis | 37 | 45 | |
| Cirrhosis | 18 | 34 | |
| Pugh-Child’s classification | 0.044* | ||
| A | 50 | 84 | |
| B | 7 | 3 | |
| Hepatitis virus | 0.757 | ||
| Absent | 9 | 18 | |
| HBV | 15 | 22 | |
| HCV | 33 | 47 | |
| AFP (ng/ml) | 0.002* | ||
| ≤ 20 | 22 | 56 | |
| > 20 | 35 | 31 | |
| PIVKA II (mAU/ml) | 0.002* | ||
| ≤ 40 | 14 | 44 | |
| > 40 | 43 | 43 | |
| Tumor multiplicity | 0.078 | ||
| Solitary | 40 | 72 | |
| Multiple | 17 | 15 | |
| Tumor size | 0.120 | ||
| < 3.0 cm | 14 | 32 | |
| ≥ 3.0 cm | 43 | 55 | |
| Differentiation | 0.009* | ||
| Well | 7 | 28 | |
| Moderate | 43 | 55 | |
| Poor | 7 | 4 | |
| Growth type | 0.495 | ||
| Expansive growth | 46 | 74 | |
| Invasive growth | 11 | 13 | |
| Serosal infiltration | 0.031* | ||
| Absent | 49 | 60 | |
| Present | 23 | 12 | |
| Formation of capsule | <0.001* | ||
| Absent | 33 | 76 | |
| Present | 24 | 11 | |
| Infiltration to capsule | 0.035* | ||
| Absent | 20 | 46 | |
| Present | 37 | 41 | |
| Septum formation | 0.036* | ||
| Absent | 14 | 36 | |
| Present | 43 | 51 | |
| Vascular invasion | <0.001* | ||
| Absent | 34 | 74 | |
| Present | 23 | 13 | |
| UICC pathological stage | <0.001* | ||
| I | 25 | 65 | |
| II | 22 | 15 | |
| III | 10 | 7 | |
Abbreviations: HBV hepatitis B virus, HCV hepatitis C virus, AFP α-fetoprotein, PIVKA protein induced by vitamin K antagonists, UICC Union for International Cancer Control. *Statistically significant difference (p < 0.05)
Fig. 3.
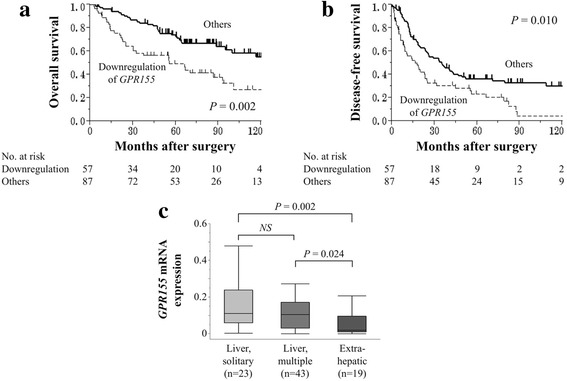
a Correlation between GPR155 expression and overall survival of patients with HCC. Overall survival of patients with downregulation of GPR155 was significantly shorter than that of patients without downregulation. b Correlation between GPR155 expression and recurrence-free survival of patients with HCC. c GPR155 mRNA levels in HCC tissues categorized by the initial recurrence pattern
We next evaluated correlations between GPR155 expression and site of the initial recurrence. The mean GPR155 expression level was significantly lower in patients who experienced extrahepatic (distant) recurrences compared with those with intrahepatic (liver confined) recurrences (Fig. 3c). Similar expression levels of GPR155 mRNA were observed in both HCC and corresponding non-cancerous tissues according to the infectious status of hepatitis viruses (Fig. 4a). Patients with downregulation of GPR155 were more likely to have a shorter overall survival than those without in patient subsets with and without HBV/HCV infection (Fig. 4b).
Fig. 4.
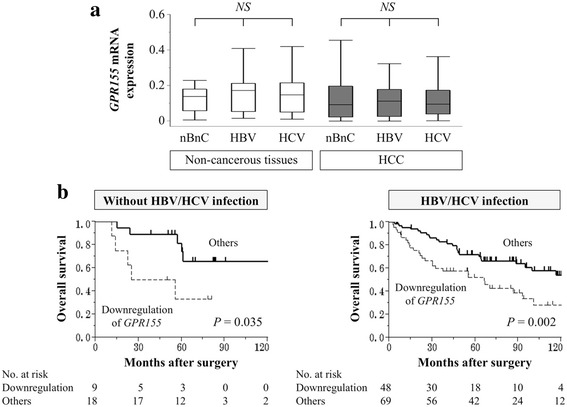
a Analysis of GPR155 mRNA expression levels according to hepatitis virus infection. b Patients with downregulation of GPR155 had significantly shorter overall survival in both the nonBnonC and HBV/HCV groups
Discussion
In the present study we evaluated the expression of GPR155 and its predictive value in HCC. The GPCR superfamily of membranous receptors, of which GPR155 is a member, has a variety of roles in intracellular signal transduction [21, 22]. When various ligands are recognized by GPCRs, GDP is converted to GTP and the α subunit and βγ subunit, acting as individual effector molecules, dissociate from the GPCR and are reported to be involved in multiple processes of cancer progression [11, 22, 23]. GPR155 harbors an auxin efflux carrier domain, a pleckstrin/G protein-interacting region, and a winged helix repressor DNA-binding domain; however, the function of the receptor is poorly understood [12, 14]. There have been some reports of GPR155 expression in mouse models, such as aberrant expression of GPR155 in UV-induced melanoma and Huntington’s disease models [12, 24]. With respect to human neoplasms, only one microarray analysis indicated suppression of GPR155 in thyroid tumor, and to our best knowledge this study is the first to evaluate GPR155 expression in digestive cancers, including HCC [14].
We found that GPR155 mRNA expression was decreased in 89% of HCC cell lines compared with the control non-tumorigenic cell line. As promoter hypermethylation is recognized as one of the prominent regulatory mechanisms of gene transcription [25], we conducted bisulfate sequence analysis to determine mechanisms of GPR155 suppression; however, no methylation was detected at the CpG island within the promoter region of GPR155 gene in any of the cell lines tested. We then performed copy number analysis to explore an alternative mechanism of GPR155 transcription because analysis of copy number variations on a genomic scale has been reported to be useful for assessing cancer progression and identifying congenital genetic abnormalities. Moreover, accumulating evidence indicates that loss of heterozygosity, mutations, and homozygous deletions are frequently present at human chromosome 2q31, the location of the GPR155 gene [26, 27]. We found copy number alterations in five (56%) HCC cell lines that showed reduced expression levels of GPR155 mRNA. These results indicated that copy number alteration might be one of the major regulatory mechanisms of GPR155 transcription. However, some HCC cell lines with decreased GPR155 mRNA expression did not show copy number alterations. When referring to The Cancer Genome Atlas database for HCC via the cBioPortal (http://www.cbioportal.org/), mutations and copy number alterations were found 0.8% and 2% of HCC tissues, respectively, though our data showed more frequent copy number alterations in HCC cell lines. Further investigation of other molecular modifications, such as acetylation of histone and microRNA expression, is expected to increase our understanding of GPR155 regulation in HCC.
In clinical samples, GPR155 mRNA levels were decreased in HCC tissues compared with the corresponding non-cancerous tissues, consistent with the results in cell lines. GPR155 mRNA expression levels were equivalent among normal liver, hepatitis, and cirrhosis as background liver status. These findings suggested that alteration of GPR155 expression may represent a specific event that occurs in the final stage of the initiation of HCC or during disease progression. Downregulation of GPR155 was associated with more aggressive phenotypes of HCC, and subsequently linked to poorer postoperative survival. GPR155 protein was successfully detected by immunohistochemical staining and we found a close correlation between GPR155 protein and mRNA expression, which allowed us to evaluate the clinical significance of GPR155 mRNA levels in a quantitative manner. Furthermore, this result may emphasize the clinical utility of GPR155 because immunohistochemical staining is a convenient and popular method commonly available in most hospitals. Both liver biopsy samples and surgically-resected specimens can be applicable in this context.
HBV and HCV infection have been recognized as major causes of HCC [2, 28]. In the latest decade, the incidence of HCV-related HCC has been dramatically declining due to increased adoption of precautions and the introduction of a direct-acting anti-HCV agent [3, 29]. Accordingly, nonBnonC-HCC arising from chronic hepatic disease, including nonalcoholic steatohepatitis and nonalcoholic fatty liver disease, is becoming increasingly important in clinical practice [7, 30–32]. In this study, we conducted a subset analysis according to hepatitis virus infection. No significant differences in GPR155 expression levels were observed among the nonBnonC, HBV, and HCV groups for both HCC and non-cancerous tissues. In previous literature it has been reported that nonBnonC-HCC is more prevalent in male patients, has relatively low transaminase levels, larger tumor size, advanced disease stage at the time of diagnosis, and a worse prognosis compared with HBV/HCV-related HCCs [7, 31, 33]. We found that the prognostic impact of GPR155 expression was equivalent in nonBnonC and HBV/HCV-related HCCs. These findings highlight the clinical utility of GPR155 expression as a prognosticator regardless of hepatitis virus infection.
Another notable finding of our study was that GPR155 expression was associated not only with overall survival but also with initial recurrence patterns. The fact that downregulation of GPR155 had a more remarkable effect on overall survival than disease-free survival motivated us to investigate the association between GPR155 expression and initial recurrence patterns. Recurrence sites represent a serious issue in the management of HCC. In cases with liver-confined recurrences, repetition of liver resection is applicable and long-term survival can be expected [2]. In contrast, the prognosis of patients with extrahepatic recurrences is dismal due to the lack of effective systemic chemotherapy [3, 9, 34]. To date, there are no biomarkers for prediction of the recurrence patterns of HCC. Our findings indicate that physicians can make a risk stratification of distant recurrences and poor prognosis by determining the expression levels of GPR155 using liver biopsies or surgical samples. Moreover, the expression levels of GPR155 may serves as a biomarker to establish a criterion for determining an appropriate therapeutic strategy such as topical therapy or systemic chemotherapy. For future consideration, external validation is necessary.
This study was limited by its lack of sufficient functional analysis of GPR155, which tempers the conclusion that it acts as a tumor suppressor in HCC. Further studies including pathway analysis and functional analysis by forced expression experiments are expected to clarify the molecular mechanisms underlying the biological activities of GPR155 in HCC.
Conclusion
Taken together, our results indicate that downregulation of GPR155 might be a prognostic factor and a predictor of initial recurrence sites, independent of hepatitis virus infection. Evaluation of GPR155 expression might improve patient follow-up and treatment after liver resection, possibly leading to better prognosis.
Additional files
Primers used in this study and annealing temperature (DOC 40 kb)
Prognostic factors for overall survival in 144 patients with hepatocellular carcinoma (DOC 49 kb)
Prognostic factors for disease-free survival in 144 patients with hepatocellular carcinoma (DOC 50 kb)
Acknowledgements
N/A.
Funding
No source of funding and material support.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- cDNA
complementary DNA
- GPCR
G protein-coupled receptor
- GPR155
G protein-coupled receptor 155
- HBV
Hepatitis B virus
- HCC
Hepatocellular carcinoma
- HCV
hepatitis C virus
- qRT-PCR
quantitative real-time reverse-transcription polymerase chain reaction
- UICC
Union for International Cancer Control
Authors’ contributions
SU, MK, HT and MH performed experiments and data analysis. HS, SY, TF, HT, YN, NI, CT, DK and MF collected cases and clinical data. MK and YK conceived and designed the study, and prepared the initial manuscript. YK supervised the project. All authors contributed to the final manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
This study conformed to the ethical guidelines of the World Medical Association Declaration of Helsinki-Ethical Principles for Medical Research Involving Human Subjects and has been approved by the Institutional Review Board of Nagoya University, Japan (No. 2013–0295). Written informed consent for usage of clinical samples and data, and publication as required by the institutional review board, was obtained from all patients.
Consent to publish
Not Applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s12885-017-3629-2) contains supplementary material, which is available to authorized users.
Contributor Information
Shinichi Umeda, Email: umeshin4018@hotmail.co.jp.
Mitsuro Kanda, Phone: +81-52-744-2249, Email: m-kanda@med.nagoya-u.ac.jp.
Hiroyuki Sugimoto, Email: sugi@med.nagoya-u.ac.jp.
Haruyoshi Tanaka, Email: tanakaharu@med.nagoya-u.ac.jp.
Masamichi Hayashi, Email: m-hayashi@med.nagoya-u.ac.jp.
Suguru Yamada, Email: suguru@med.nagoya-u.ac.jp.
Tsutomu Fujii, Email: fjt@med.nagoya-u.ac.jp.
Hideki Takami, Email: takamihideki@med.nagoya-u.ac.jp.
Yukiko Niwa, Email: yukiko-niwa@med.nagoya-u.ac.jp.
Naoki Iwata, Email: iwt-nk@med.nagoya-u.ac.jp.
Chie Tanaka, Email: chtanaka@med.nagoya-u.ac.jp.
Daisuke Kobayashi, Email: kobadai@med.nagoya-u.ac.jp.
Michitaka Fujiwara, Email: mfuji@med.nagoya-u.ac.jp.
Yasuhiro Kodera, Email: ykodera@med.nagoya-u.ac.jp.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet. 2012;379:1245–1255. doi: 10.1016/S0140-6736(11)61347-0. [DOI] [PubMed] [Google Scholar]
- 3.Poon D, Anderson BO, Chen LT, Tanaka K, Lau WY, Van Cutsem E, Singh H, Chow WC, Ooi LL, Chow P, Khin MW, Koo WH. Management of hepatocellular carcinoma in Asia: consensus statement from the Asian oncology summit 2009. Lancet Oncol. 2009;10:1111–1118. doi: 10.1016/S1470-2045(09)70241-4. [DOI] [PubMed] [Google Scholar]
- 4.Ezaka K, Kanda M, Sugimoto H, Shimizu D, Oya H, Nomoto S, Sueoka S, Tanaka Y, Takami H, Hashimoto R, Okamura Y, Yamada S, Fujii T, Nakayama G, Koike M, Fujiwara M, Kodera Y. Reduced expression of Adherens junctions associated protein 1 predicts recurrence of hepatocellular carcinoma after curative hepatectomy. Ann Surg Oncol. 2015;22(Suppl 3):1499–1507. doi: 10.1245/s10434-015-4695-9. [DOI] [PubMed] [Google Scholar]
- 5.Kanda M, Sugimoto H, Kodera Y. Genetic and epigenetic aspects of initiation and progression of hepatocellular carcinoma. World J Gastroenterol. 2015;21:10584–10597. doi: 10.3748/wjg.v21.i37.10584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sobin LH, Gospodarowicz MK, C W: International Union against Cancer, TNM classification of malignant tumors. Seventh edition. New York: Wiley-Blackwell 2009.
- 7.Bruix J, Gores GJ, Mazzaferro V. Hepatocellular carcinoma: clinical frontiers and perspectives. Gut. 2014;63:844–855. doi: 10.1136/gutjnl-2013-306627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sueoka S, Kanda M, Sugimoto H, Shimizu D, Nomoto S, Oya H, Takami H, Ezaka K, Hashimoto R, Tanaka Y, Okamura Y, Yamada S, Fujii T, Nakayama G, Koike M, Fujiwara M, Kodera Y. Suppression of SAMSN1 expression is associated with the malignant phenotype of hepatocellular carcinoma. Ann Surg Oncol. 2015;22(Suppl 3):1453–1460. doi: 10.1245/s10434-015-4524-1. [DOI] [PubMed] [Google Scholar]
- 9.El-Serag HB. Hepatocellular carcinoma. N Engl J Med. 2011;365:1118–1127. doi: 10.1056/NEJMra1001683. [DOI] [PubMed] [Google Scholar]
- 10.Yang JD, Roberts LR. Hepatocellular carcinoma: A global view. Nat Rev Gastroenterol Hepatol. 2010;7:448–458. doi: 10.1038/nrgastro.2010.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Venkatakrishnan AJ, Deupi X, Lebon G, Heydenreich FM, Flock T, Miljus T, Balaji S, Bouvier M, Veprintsev DB, Tate CG, Schertler GF, Babu MM. Diverse activation pathways in class a GPCRs converge near the G-protein-coupling region. Nature. 2016;536:484–7. [DOI] [PMC free article] [PubMed]
- 12.Trifonov S, Houtani T, Shimizu J, Hamada S, Kase M, Maruyama M, Sugimoto T. GPR155: gene organization, multiple mRNA splice variants and expression in mouse central nervous system. Biochem Biophys Res Commun. 2010;398:19–25. doi: 10.1016/j.bbrc.2010.05.162. [DOI] [PubMed] [Google Scholar]
- 13.Shimizu D, Kanda M, Tanaka H, Kobayashi D, Tanaka C, Hayashi M, Iwata N, Niwa Y, Takami H, Yamada S, Fujii T, Nakayama G, Fujiwara M, Kodera Y. GPR155 serves as a predictive biomarker for Hematogenous metastasis in patients with gastric cancer. Sci Rep. 2017;7 doi: 10.1038/srep42089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schulten HJ, Al-Mansouri Z, Baghallab I, Bagatian N, Subhi O, Karim S, Al-Aradati H, Al-Mutawa A, Johary A, Meccawy AA, Al-Ghamdi K, Al-Hamour O, Al-Qahtani MH, Al-Maghrabi J. Comparison of microarray expression profiles between follicular variant of papillary thyroid carcinomas and follicular adenomas of the thyroid. BMC Genomics. 2015;1(16 Suppl):S7. doi: 10.1186/1471-2164-16-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oya H, Kanda M, Sugimoto H, Shimizu D, Takami H, Hibino S, Hashimoto R, Okamura Y, Yamada S, Fujii T, Nakayama G, Koike M, Nomoto S, Fujiwara M, Kodera Y. Dihydropyrimidinase-like 3 is a putative hepatocellular carcinoma tumor suppressor. J Gastroenterol. 2015;50:590–600. doi: 10.1007/s00535-014-0993-4. [DOI] [PubMed] [Google Scholar]
- 16.Kanda M, Shimizu D, Tanaka H, Tanaka C, Kobayashi D, Hayashi M, Iwata N, Niwa Y, Yamada S, Fujii T, Sugimoto H, Murotani K, Fujiwara M, Kodera Y. Significance of SYT8 for the detection, prediction, and treatment of peritoneal metastasis from gastric cancer. Ann Surg. 2016. [Epub ahead of print]. doi:10.1097/SLA.0000000000002096. [DOI] [PubMed]
- 17.Kanda M, Shimizu D, Fujii T, Tanaka H, Tanaka Y, Ezaka K, Shibata M, Takami H, Hashimoto R, Sueoka S, Iwata N, Kobayashi D, Tanaka C, Yamada S, Nakayama G, Sugimoto H, Koike M, Fujiwara M, Kodera Y. Neurotrophin receptor-interacting melanoma antigen-encoding gene homolog is associated with malignant phenotype of gastric cancer. Ann Surg Oncol. 2016;23(Suppl 4):532–9. [DOI] [PubMed]
- 18.Kanda M, Shimizu D, Tanaka H, Shibata M, Iwata N, Hayashi M, Kobayashi D, Tanaka C, Yamada S, Fujii T, Nakayama G, Sugimoto H, Koike M, Fujiwara M, Kodera Y. Metastatic pathway-specific transcriptome analysis identifies MFSD4 as a putative tumor suppressor and biomarker for hepatic metastasis in patients with gastric cancer. Oncotarget. 2016;7:13667–13679. doi: 10.18632/oncotarget.7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanda M, Tanaka C, Kobayashi D, Tanaka H, Shimizu D, Shibata M, Takami H, Hayashi M, Iwata N, Niwa Y, Yamada S, Fujii T, Nakayama G, Fujiwara M, Kodera Y. Epigenetic suppression of the immunoregulator MZB1 is associated with the malignant phenotype of gastric cancer. Int J Cancer. 2016;139:2290–2298. doi: 10.1002/ijc.30286. [DOI] [PubMed] [Google Scholar]
- 20.Kanda M, Shimizu D, Fujii T, Sueoka S, Tanaka Y, Ezaka K, Takami H, Tanaka H, Hashimoto R, Iwata N, Kobayashi D, Tanaka C, Yamada S, Nakayama G, Sugimoto H, Koike M, Fujiwara M, Kodera Y. Function and diagnostic value of Anosmin-1 in gastric cancer progression. Int J Cancer. 2016;138:721–730. doi: 10.1002/ijc.29803. [DOI] [PubMed] [Google Scholar]
- 21.Beautrait A, Michalski KR, Lopez TS, Mannix KM, McDonald DJ, Cutter AR, Medina CB, Hebert AM, Francis CJ, Bouvier M, Tesmer JJ, Sterne-Marr R. Mapping the putative G protein-coupled receptor (GPCR) docking site on GPCR kinase 2: insights from intact cell phosphorylation and recruitment assays. J Biol Chem. 2014;289:25262–25275. doi: 10.1074/jbc.M114.593178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Purvanov V, Holst M, Khan J, Baarlink C, Grosse R. G-protein-coupled receptor signaling and polarized actin dynamics drive cell-in-cell invasion. elife. 2014;3:e02786. [DOI] [PMC free article] [PubMed]
- 23.Meiri D, Marshall CB, Mokady D, LaRose J, Mullin M, Gingras AC, Ikura M, Rottapel R. Mechanistic insight into GPCR-mediated activation of the microtubule-associated RhoA exchange factor GEF-H1. Nat Commun. 2014;5:4857. doi: 10.1038/ncomms5857. [DOI] [PubMed] [Google Scholar]
- 24.Hacker E, Muller K, Whiteman DC, Pavey S, Hayward N, Walker G. Reduced expression of IL-18 is a marker of ultraviolet radiation-induced melanomas. Int J Cancer. 2008;123:227–231. doi: 10.1002/ijc.23389. [DOI] [PubMed] [Google Scholar]
- 25.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 26.Li SP, Wang HY, Li JQ, Zhang CQ, Feng QS, Huang P, XJ Y, Huang LX, Liang QW, Zeng YX. Genome-wide analyses on loss of heterozygosity in hepatocellular carcinoma in southern China. J Hepatol. 2001;34:840–849. doi: 10.1016/S0168-8278(01)00047-2. [DOI] [PubMed] [Google Scholar]
- 27.Nishimura T, Nishida N, Komeda T, Fukuda Y, Nakao K. Genotype stability and clonal evolution of hepatocellular carcinoma assessed by autopsy-based genome-wide microsatellite analysis. Cancer Genet Cytogenet. 2005;161:164–169. doi: 10.1016/j.cancergencyto.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 28.El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–2576. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- 29.Webster DP, Klenerman P, Dusheiko GM. Hepatitis C. Lancet. 2015;385:1124–1135. doi: 10.1016/S0140-6736(14)62401-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ahmed A, Wong RJ, Harrison SA. Nonalcoholic fatty liver disease review: diagnosis, treatment, and outcomes. Clin Gastroenterol Hepatol. 2015;13:2062–2070. doi: 10.1016/j.cgh.2015.07.029. [DOI] [PubMed] [Google Scholar]
- 31.Charrez B, Qiao L, Hebbard L. Hepatocellular carcinoma and non-alcoholic steatohepatitis: the state of play. World J Gastroenterol. 2016;22:2494–2502. doi: 10.3748/wjg.v22.i8.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eslam M, George J. Genetic and epigenetic mechanisms of NASH. Hepatol Int. 2016;10:394–406. doi: 10.1007/s12072-015-9689-y. [DOI] [PubMed] [Google Scholar]
- 33.Yki-Jarvinen H. Non-alcoholic fatty liver disease as a cause and a consequence of metabolic syndrome. Lancet Diabetes Endocrinol. 2014;2:901–910. doi: 10.1016/S2213-8587(14)70032-4. [DOI] [PubMed] [Google Scholar]
- 34.Sanyal AJ, Yoon SK, Lencioni R. The etiology of hepatocellular carcinoma and consequences for treatment. Oncologist. 2010;15(Suppl 4):14–22. doi: 10.1634/theoncologist.2010-S4-14. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primers used in this study and annealing temperature (DOC 40 kb)
Prognostic factors for overall survival in 144 patients with hepatocellular carcinoma (DOC 49 kb)
Prognostic factors for disease-free survival in 144 patients with hepatocellular carcinoma (DOC 50 kb)
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.


