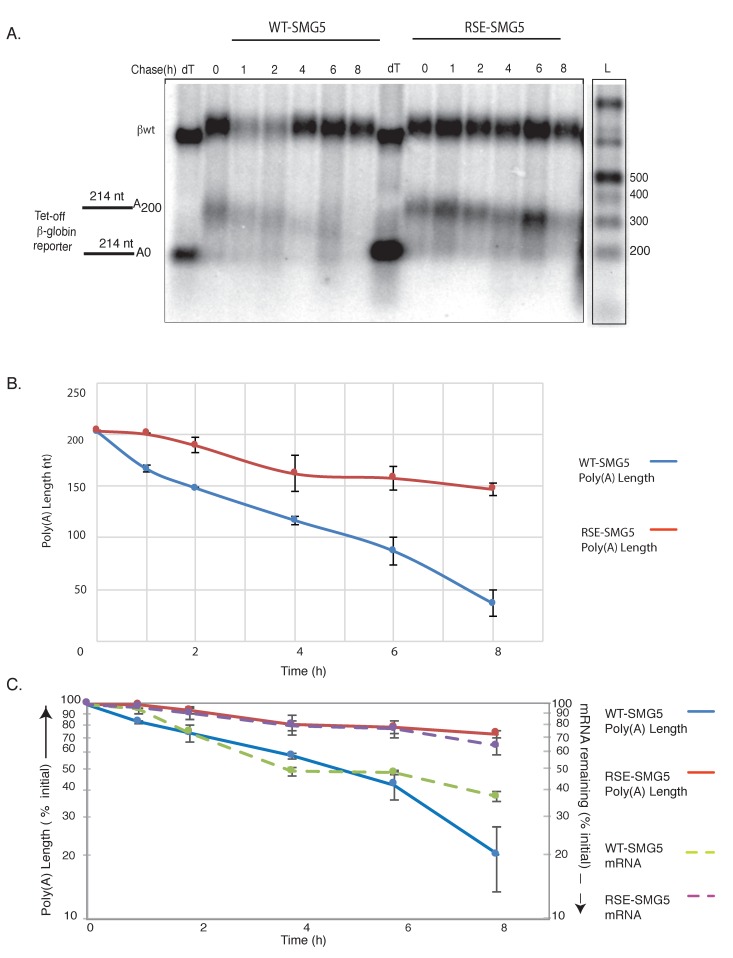
Figure 2.
RNA stability element inhibits deadenylation. (A) Agarose northern blot showing deadenylation and decay of WT-SMG5 and RSE-SMG5 mRNAs following tet-off transcriptional pulse chase. RNA was subjected to oligonucleotide-directed RNaseH treatment prior to Northern blotting for tail length determination. Lanes marked “dT” show the length of the RNA fragment without a poly(A) tail; 1 kb plus DNA ladder (ThermoFisher Catalog number: 10787018, Halethorpe, MD, USA) was end labelled and used for size determination (size corrected for RNA). Wildtype β-globin (βwt) (upper band) was used as a loading control. (B) Poly(A) tail length determined from the gel was plotted against time to show the initial and final tail length of the two reporter constructs. The plotted values are the average of at least three separate experiments. Error bars show the standard error. Deadenylation rate of the reporters were also determined from this analysis. (C) Deadenylation and decay of the reporter mRNAs were compared by plotting mRNA remaining (% of original) and poly(A) length remaining (% of original tail length) on the same plot.