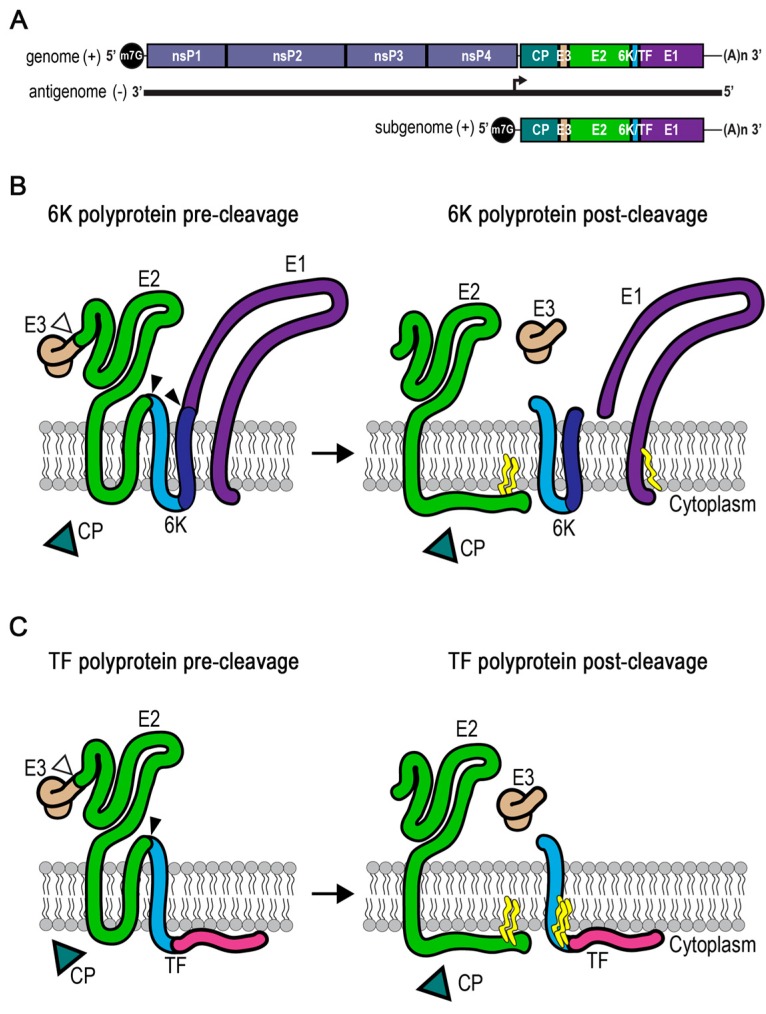
Figure 1.
Structural polyprotein membrane topology and processing. (A) The positive-sense RNA alphavirus genome has two open reading frames. At the 5’ end is the methyl-7-guanosine cap (m7G), followed by the nonstructural protein (nsP) and structural protein open reading frames, and the 3’ poly A tail. Nonstructural proteins are translated directly from the genome. During replication, the minus-sense antigenome is used as the template for producing the subgenomic RNA from an internal subgenomic promoter (arrow). Structural proteins are translated from the subgenomic transcript. (B,C) Pre-cleavage structural polyprotein conformation in the endoplasmic reticulum (ER) membrane. Capsid (CP) is first released as a soluble protein in the cytoplasm. The N-terminus of E3 contains a signal sequence that directs translocation across the membrane. Open arrows mark the furin cleavage site between E3 and E2, which occurs in the trans-Golgi network. The remaining envelope proteins are threaded through the membrane with insertion of transmembrane domains. Closed arrows mark signalase cleavage sites that are used in the ER. Post-cleavage conformations include palmitoylation sites (yellow). (B) The 6K polyprotein is the majority translation product. (C) The TF form of the polyprotein is the minority product resulting from frameshifting in the 6K gene, and does not include E1.