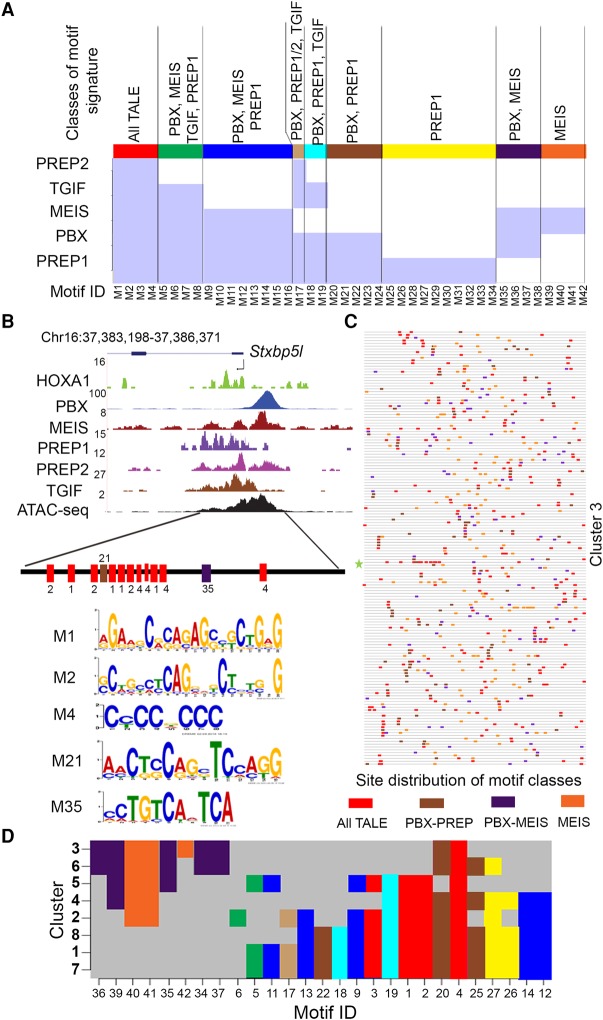
Figure 3.
Unique and overlapping genome-wide binding preferences of individual TALE proteins and their distribution in HOXA1-bound regions. (A) MEME and AME (analysis of motif enrichment) identify motifs in genome-wide binding preferences for members of the TALE family (PBX, MEIS, TGIF, PREP1, and PREP2). There are 42 enriched motif signatures (M1–M42) comprised of both novel and known consensus TALE binding sites. These 42 cis-motifs are divided into nine distinct classes based on their presence or absence within regions bound by one or more of the TALE family members (shaded light blue areas in the matrix). Each class is identified by a separate color (top) along with the respective TALE family members showing occupancy on these motifs. (B) UCSC Genome Browser shot (mm10) showing occupancy of HOXA1 and TALE factors near the Stxbp5l locus (a representative binding region in Cluster 3). The diagram at the bottom illustrates the five different motifs (M1, M2, M4, M21, and M35) and represents three classes of motif signatures defined in A, above. There are multiple copies of three of the cis-motifs (M1, M2, and M4). (C) A block diagram shows the occurrence of four distinct motif classes with multiple copies in all the loci present in Cluster 3. Each respective motif class is colored as indicated in A. Length of peaks are not drawn to scale. (D) A matrix indicating the presence or absence of the individual classes of cis-motif signatures in the respective clusters defined by combinatorial TALE binding on HOXA1-bound regions. Distribution of various motif signature classes in each cluster are colored as defined in A. This plots the unique combinations of motif signatures which underlie the differential binding of TALE proteins in each cluster.