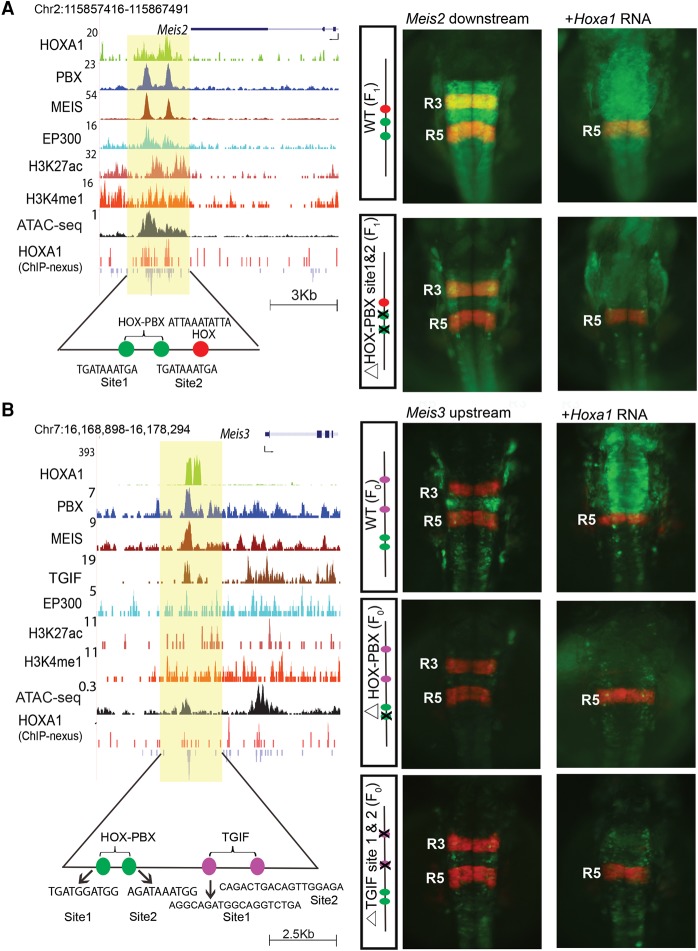
Figure 4.
The Meis2 and Meis3 genes are downstream targets of HOXA1. UCSC Genome Browser shots of Meis2 (A) and Meis3 (B) show HOXA1-bound regions along with occupancy of PBX and MEIS, activator protein EP300, and open chromatin. For Meis3, occupancy of TGIF is also shown. ChIP-nexus data reveal the presence of multiple binding motifs for HOXA1 and TALE cofactors in the protected regions as noted at the bottom. The shaded area indicates bound regions. Panels to the right are transgenic zebrafish reporter images demonstrating the regulatory potential of the binding regions ±250 bp. For both Meis2 and Meis3, transgenic embryos show restricted reporter expression in the hindbrain and a response to ectopic expression of Hoxa1, as indicated by the loss of RFP (red)-specific expression in R3 and expansion of GFP (green) expression. In both cases, deletion of the HOX-PBX sites abolishes or reduces GFP reporter activity and reduces response to ectopic Hoxa1 mRNA. For Meis3 (B), mutation of the two TGIF motifs in the enhancer result in low expression of reporter and weak anterior expansion upon coinjection with Hoxa1 mRNA. HOX-PBX bipartite motifs and TGIF motifs in each of the enhancers are shown at the bottom. All embryos are shown in dorsal views. (F0) Founder embryos, (F1) stable lines carrying the reporter.