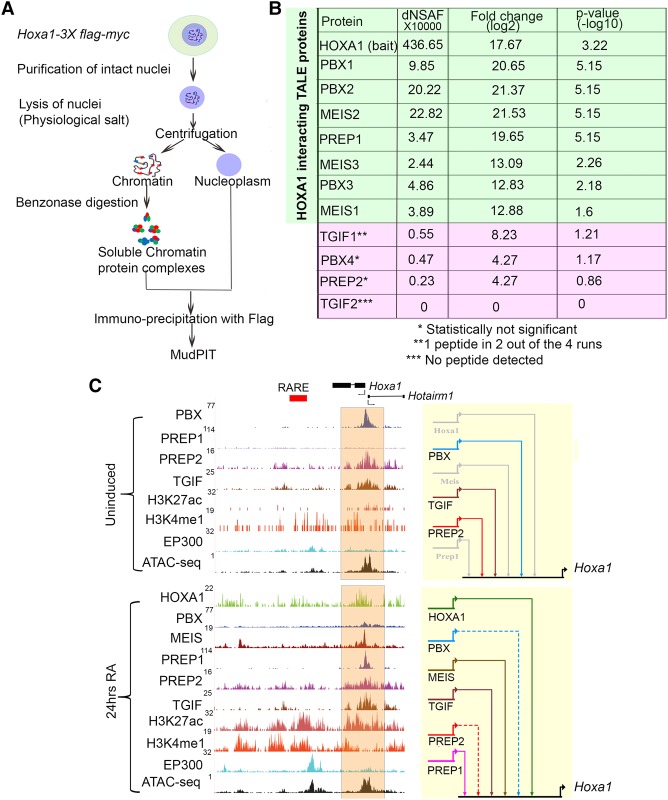
Figure 5.
Interactions between HOXA1 and TALE proteins on chromatin and binding of TALE proteins to the Hoxa1 locus. (A) Strategy for isolation and identification of chromatin-bound complexes interacting with HOXA1 by immunoprecipitation and MudPIT. (B) MudPIT analyses averaged from four replicates showing dNSAF(X10000), fold enrichment, and P-values of TALE proteins associated with HOXA1 on chromatin. (C) UCSC Genome Browser shots (mm10) for the Hoxa1 locus. Binding of HOXA1 and occupancy of the TALE family members, modified histone marks (H3K4me1 and H3K27Ac) characteristic of enhancers, occupancy of the EP300 activator protein, and accessibility of chromatin (ATAC-seq) are shown in uninduced (top) and differentiated (bottom) cells. The orange shaded area indicates Hoxa1-bound regions near the promoter. In the BioTapestry plots (right panels), solid colored lines indicate binding of each specific TALE factor, while gray lines indicate lack of binding. Dotted lines represent decreased binding.