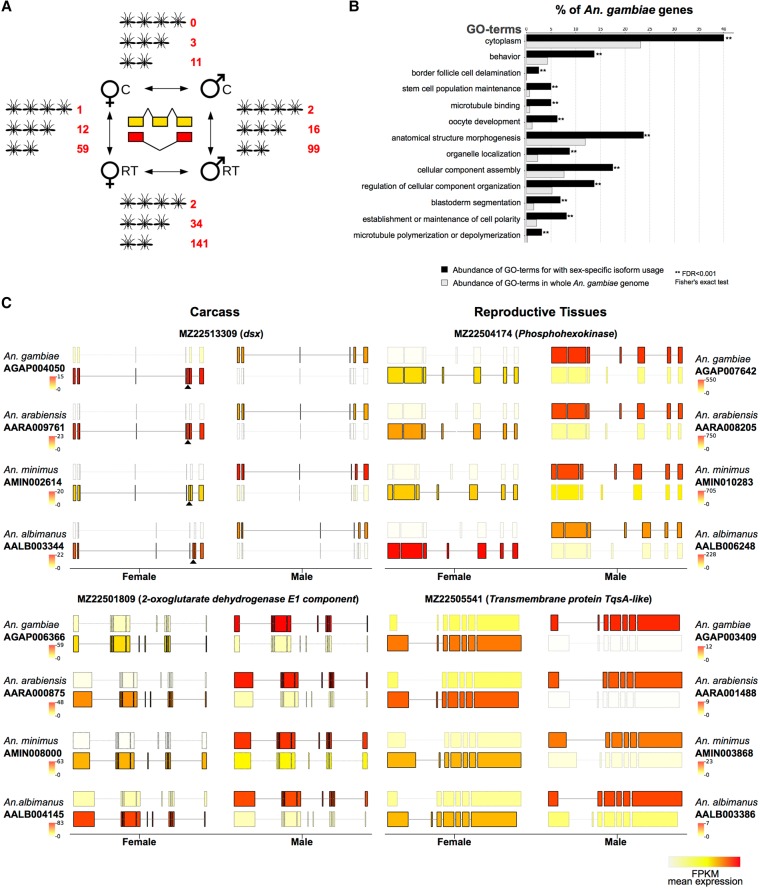
Figure 6.
Sex- and tissue-specific differential transcript isoform usage. (A) Overview of the number of orthologs displaying conserved alternative isoform usage conserved in four, three, or two species, respectively. (B) Gene Ontology functional enrichment analysis showing enriched GO-terms in An. gambiae among genes displaying sex-specific isoform usage, compared to the entire genome. (C) Two examples of sex-specific isoform usage between males and females are shown for carcass (left) and reproductive tissues (right). In each case, only the predominant isoforms in the two sexes are shown, with the color of the transcript displaying transcript expression levels (in FPKM). The female-specific exon of the well-characterized doublesex (dsx) locus is marked with a black triangle. Although visual inspection of splicing pattern highlights consistent use of alternative sex-specific splicing in all four species, the dsx locus passed the cutoffs only in three species. A similar pattern was also observed for a number of other genes and suggests that this analysis is stringent.