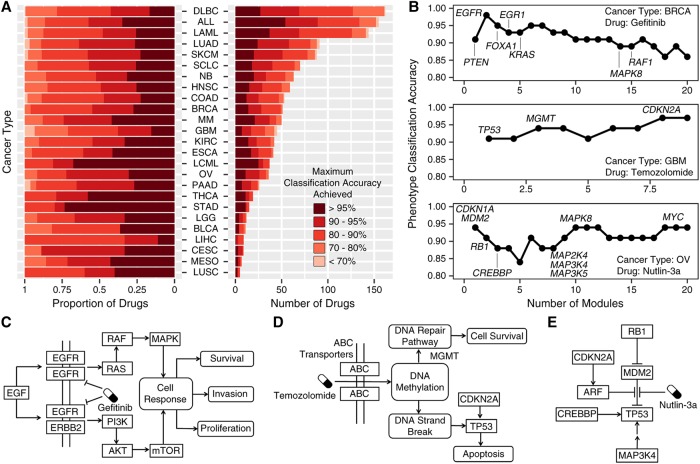
Figure 6.
Drug efficacy predicted by HIT'nDRIVE-seeded driver genes. (A) Accuracy of drug-response phenotype classification for all 265 drugs used in the GDSC study across 25 cancer types (the remaining five cancer types for which only a very limited number of cell lines have been made available are statistically insignificant and thus have not been used). The classification accuracy for each drug on each cancer type is measured based on the collective use of at most 10 best-discriminating modules; i.e., the accuracy is maximized across the range of one to 10 (best-discriminating) modules. Note that many of the drugs were not tested on all cancer types; in fact, for the vast majority of cancer types, only a handful of drugs were tested. (B) Classification accuracy of modules that distinguish the drug-response phenotypes after treatment with Gefitinib in BRCA cell lines (top), temozolomide in GBM cell lines (middle), and Nutlin-3a in OV cell lines (bottom). Important genes identified in the modules and involved in the dysregulated signaling pathways have been highlighted. (C–E) The figures represent the dysregulated signaling pathways in the respective drug perturbation.