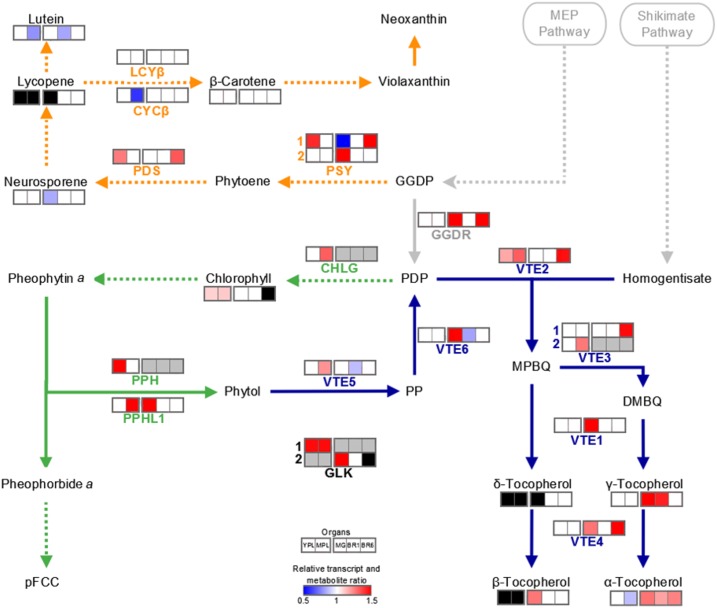
Figure 10.
Isoprenoid metabolism. Schematic representation shows the interconnection between chlorophyll synthesis and degradation (green), carotenogenesis (orange), and tocopherol biosynthetic (blue) pathways. Dotted lines denote that intermediate steps were omitted. The heat map represents statistically significant differences in relative transcript and metabolite amounts detected in SlORE1S02 knockdown lines compared with the corresponding organ of the MT control genotype (P < 0.05). For simplicity, the mean of three transgenic line values is represented when at least two were statistically significant different compared with the MT control. The absolute metabolite and relative transcript values are detailed in Supplemental Tables S3 and S4. Black and gray colors indicate that transcripts were not detected or not addressed, respectively. Enzymes and compounds are named according to the following abbreviations: CHLG, chlorophyll synthase; CYCβ, chromoplast-specific β-lycopene cyclase; GGDP, geranylgeranyl diphosphate; GGDR, geranylgeranyl diphosphate reductase; LCYβ, chloroplast-specific β-lycopene cyclase; MEP, methylerythritol 4-phosphate; MPBQ, 2-methyl-6-geranylgeranylbenzoquinol; PDP, phytyl diphosphate; PDS, phytoene desaturase; pFCC, primary fluorescent chlorophyll catabolite; PP, phytyl phosphate; PPH, pheophytinase; PPHL1, pheophytinase-like1; PSY, phytoene synthase; VTE1, tocopherol cyclase; VTE2, homogentisate phytyl transferase; VTE3, 2,3-dimethyl-5-phytylquinol methyltransferase; VTE4, tocopherol C-methyl transferase; VTE5, phytol kinase; VTE6, phytyl phosphate kinase. Adapted from Almeida et al. (2016).