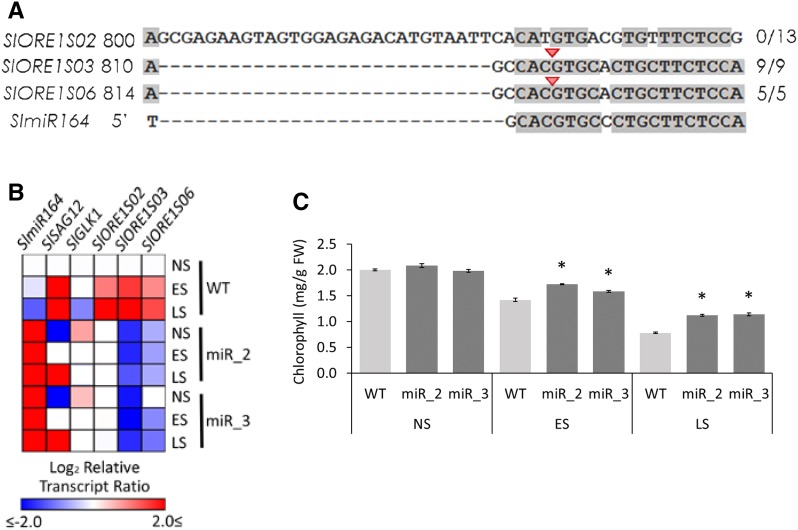
Figure 4.
Regulation of SlORE1s by SlmiR164. A, Highlight of the alignment of SlORE1s and SlmiR164 showing the putative binding sites. Validation by a modified 5′ RACE assay identified cleaved transcripts of SlORE1S03 and SlORE1S06. The red arrowheads and the numbers on the right indicate the inferred cleavage sites and the fractions of positive cloned PCR products ending at the site, respectively. The shading threshold of alignment = 75%. B, Heat map representing the transcript profiles of nonsenescent (NS), early-senescing (ES), and late-senescing (LS) leaves of wild-type control (WT) and OEmiR164a (lines miR_2 and miR_3) genotypes. Values represent means of at least three biological replicates normalized against the nonsenescent wild-type sample. Statistically significant differences in comparison with the nonsenescent wild type are represented as colored squares (P < 0.05). The relative transcript values are detailed in Supplemental Table S2. C, Chlorophyll contents in NS, ES, and LS leaves. Values represent means ± se from at least three biological replicates. Asterisks denote statistically significant differences compared with the wild-type control (P < 0.05.) FW, Fresh weight.