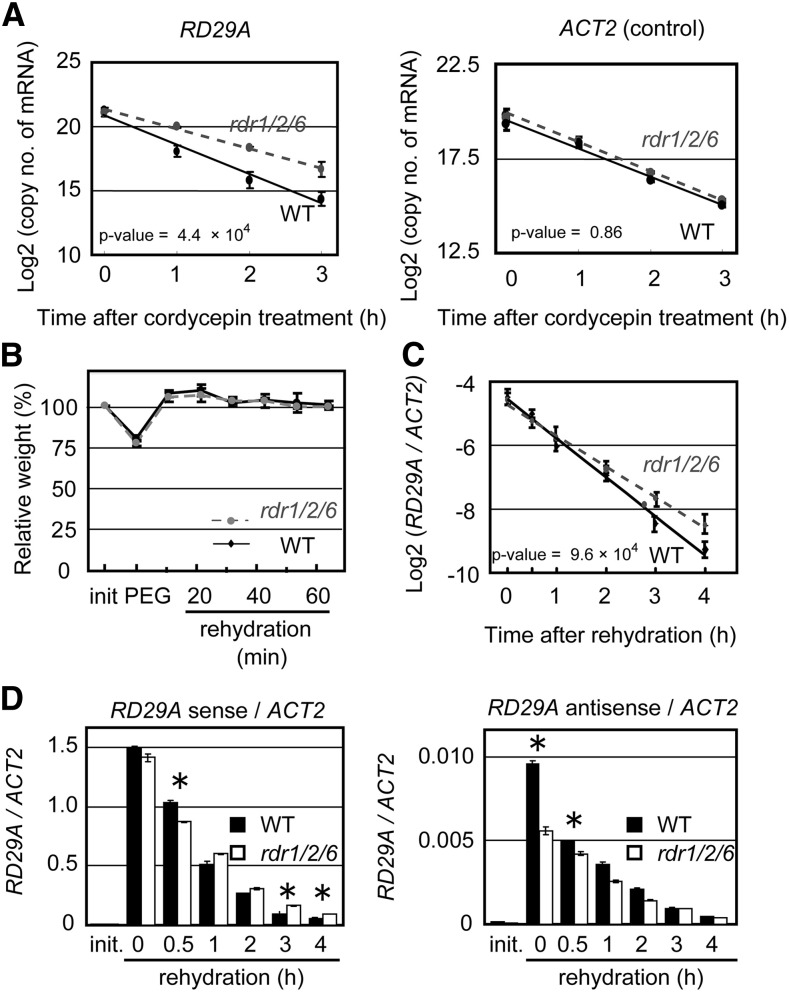
Figure 5.
RNA Decay Rate of RD29A Poly (A+) Sense RNA in wild-type and rdr1/2/6 plants subjected to an osmotic stress. A, Two-week-old wild-type and rdr1/2/6-1 plants were pretreated with a 30% PEG solution for 2 h and then treated with 0.6 mm cordycepin, a transcription inhibitor. The RNA decay rate of RD29A (a RDR1/2/6-dependent antisense RNA locus) and ACT2 (a locus without antisense RNA) mRNAs was measured by qRT-PCR. The vertical axis indicates the log2 copy numbers of mRNA per 100 ng of total RNA. The horizontal axis indicates the time-course following cordycepin treatment. Covariance analysis (ANCOVA) was used to determine whether there are different RNA decay rates between wild-type and rdr1/2/6-1 plants. B, rdr1/2/6-1 and wild-type plants were treated with 30% PEG6000 for 2 h and returned to a well-watered condition (rehydration). Relative fresh weight of rdr1/2/6-1 and wild-type plants were then measured. Covariance analysis (ANCOVA) was used to determine whether there are different on the RNA decay rates between wild type and rdr1/2/6-1 plants. C, Accumulation of RD29A mRNA during the rehydration phase was measured by qRT-PCR and normalized to ACT2 accumulation. D, Accumulation of RD29A mRNA and antisense RNA (fAsRD29A1) during the rehydration phase was measured by qRT-PCR and normalized to ACT2 accumulation. Asterisk indicates the P values for t test < 0.05.