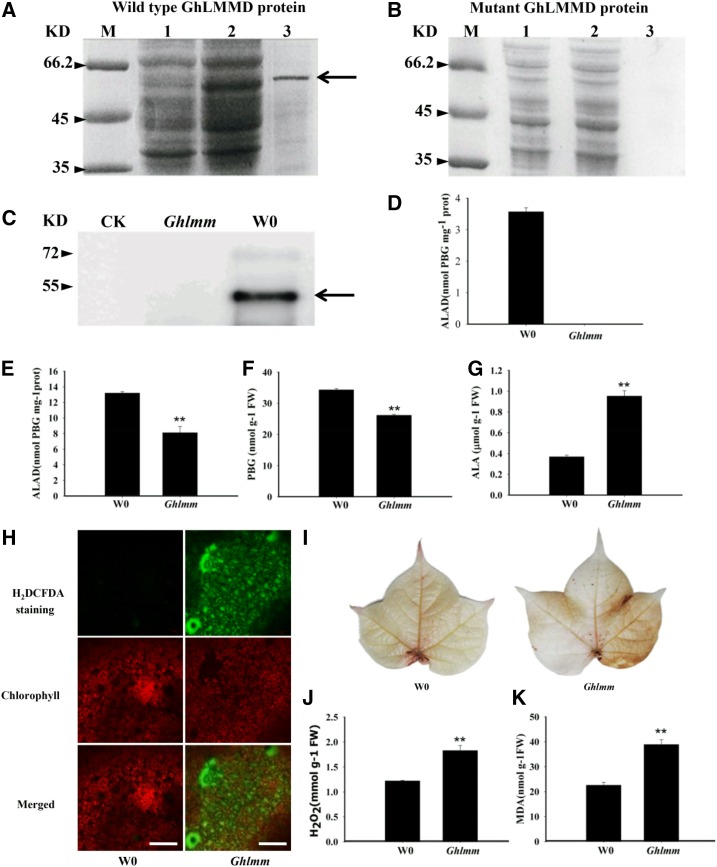
Figure 3.
GhLMMD loss of function leads to accumulation of ALA and ROS burst. A and B, Expression and purification of wild-type and mutant GhLMMD proteins in E. coli. Lane M: protein marker; Lane 1: extracts from noninduced cells; Lane 2: extracts from IPTG-induced cells; Lane 3: GhLMMD protein purified from extracts of IPTG-induced E. coli cells. Arrow indicates the induced and purified GhLMMD protein, which is the same size with the predicted protein. Note that the wild-type GhLMMD protein could be expressed and purified from E. coli, while no mutant GhLMMD protein was expressed after induction. C, Confirmation of purified ALAD protein by western blotting. Antibodies against His-tag were used in the assay. CK, Ghlmm, and W0 indicate proteins purified from E. coli transformed with empty, mutant GhLMMD, and wild-type GhLMMD vector, respectively. D, ALAD enzyme activities of purified wild-type and mutant GhLMMD proteins. E, Analysis of ALAD enzyme activities in wild-type and mutant cotton leaves. F and G, Measurements of the contents of PBG and ALA in cotton leaves. Leaves of cotton seedlings at two-true-leaves stage were used for the analysis. H, Detection of ROS levels in cotton leaves by H2DCFDA staining. Then 10 μm 2’,7’-dichlorodihydrofluorescein diacetate (H2DCFD), a ROS-specific fluorescent indicator, was used for the staining. Bars = 50 μm. I, H2O2 visualization in cotton leaves by staining with DAB, showing more H2O2 was produced in the mutant than the wild type. J and K, Measurements of the contents of H2O2 and MDA in cotton leaves. Leaves of cotton seedlings at two-true-leaves stage were used for the analysis. At least three biological replicates were performed in each experiment. *P < 0.05 and **P < 0.01 (Student’s t test).