Abstract
Transposons are a class of selfish DNA elements that can mobilize within a genome. If mobilization is accompanied by an increase in copy number (replicative transposition), the transposon may sweep through a population until it is fixed in all of its interbreeding members. This introgression has been proposed as the basis for drive systems to move genes with desirable phenotypes into target species. One such application would be to use them to move a gene conferring resistance to malaria parasites throughout a population of vector mosquitos. We assessed the feasibility of using the piggyBac transposon as a gene-drive mechanism to distribute anti-malarial transgenes in populations of the malaria vector, Anopheles stephensi. We designed synthetic gene constructs that express the piggyBac transposase in the female germline using the control DNA of the An. stephensi nanos orthologous gene linked to marker genes to monitor inheritance. Two remobilization events were observed with a frequency of one every 23 generations, a rate far below what would be useful to drive anti-pathogen transgenes into wild mosquito populations. We discuss the possibility of optimizing this system and the impetus to do so.
Keywords: Gene-drive, Genetically-engineered mosquitoes, Malaria vector
1. Introduction
Transposable elements have been proposed as a mechanistic basis for synthetic autonomous (self-mobilizing) gene-drive systems for introgressing anti-pathogen effector genes into wild mosquito populations (Kidwell and Ribeiro, 1992; Ribeiro and Kidwell, 1994; James, 2005). To function optimally, such a system must have the ability to remobilize (excise and integrate) itself and the desirable genes it is carrying from an initial insertion site to a new location in the genome of the target species. Furthermore, it should remobilize replicatively and with a frequency that allows it to reach fixation in a population within a useful timeframe (James, 2005). A piggyBac transposon construct lacking a source of transposase and integrated into the genome of the vector mosquito, Anopheles stephensi, could be remobilized to a new location by crossing the transgenic line with one expressing the transposase (O’Brochta et al., 2011). This ‘jumpstarter’ finding provided conceptual support for the development of synthetic elements capable of autonomous remobilization. We designed an autonomous gene-drive system based on the piggyBac element and the nanos gene 5′-and 3′-end flanking control DNA and tested it in transgenic An. stephensi. The construct was able to remobilize, however, at a frequency too low to be of practical use. Modifications including the replacement of control elements that promote a more robust expression of the piggyBac transposase, the use of alternative transposable elements, and the initial insertion of constructs at different locations on the mosquito genome may be able to increase remobilization efficiency.
2. Methods
2.1. Plasmids
The high-fidelity Phusion (Finnzymes, Wolburn, MA) DNA polymerase was used to amplify DNA fragments for plasmid construction. All fragments were amplified with oligonucleotide primers (Supplemental Table 1) designed with restriction sites for directional cloning into the shuttle vector pSLfa 1180fa (pSLfa; Horn and Wimmer, 2000).
2.1.1. pBac3XP3-GFP[0.9nanos-pBacORF]
Specific primers were used to amplify nanos fragments from genomic An. stephensi DNA and the piggyBac open reading frame (ORF) from a piggyBac Helper plasmid (Handler et al., 1998). Amplification products were sub-cloned first into the Zero Blunt Topo vector (Invitrogen), then sequenced and sub-cloned into pSLfa with specific enzymes: HindIII/XbaI for the 900 base-pair (bp) promoter/5′-end untranslated region (UTR), XbaI/BamHI for the 1789bp piggyBac transposase ORF and BamHI/EcoRI for the nanos 3′UTR This cassette, [0.9nanos-pBacORF], was excised from pSLfa using AscI and sub-cloned into pBac 3XP3-EGFPafm. The resulting vector, pBac3XP3-EGFP[0.9nanos-pBacORF] was used to generate the transgenic An. stephensi line As28+.
2.1.2. pBacDsRed-attB[3.8nanos-pBacORF]
The piggyBac right inverted terminal repeat (ITR) and the 3XP3-EGFP-SV40 expression cassette in the pBac3XP3-EGFP[0.9nanos-pBacORF] construct were replaced with the right ITR and the 3XP3-DsRed-SV40 expression cassette from pBac[3xP3-DsRedaf] (Nimmo et al., 2006) through the unique KasI (vector backbone) and NotI (3′-end of SV40) sites in both constructs to produce the pBac3XP3-DsRed-SV40[0.9nanos-pBacORF] plasmid.
The larger 3.8 nanos promoter sequence was synthesized de novo by Epoch Biolabs (Houston, TX) based on the genome sequence of the An. stephensi nanos gene (AY738090, ASTEI02887, Calvo et al., 2005), with XhoI and FseI sites added to the 5′-end and a HindIII site added at the 3′-end. XhoI and FseI were used to cut and clone the synthesis product into the pBluescript SK (−) plasmid to produce pBSK-Nan3.8. The larger promoter then was cut from the pBSK-Nan3.8 plasmid and used to replace the 0.9 nanos promoter in pBac3XP3-dsRed-SV40[0.9nanos-pBacORF] using the FseI and PpuMI to produce pBac3XP3-DsRed-SV40[3.8nanos-pBacORF]. The attB sequence in pBattB[3xP3-DsRed2-SV40] (Labbé et al., 2010) was amplified using attB FOR and attB REV primers (Supplemental Table 1), which incorporate a PstI site at each terminus of the product.
The DNA segment containing the PstI site in the DsRed ORF was removed by digesting pBac[3xP3-DsRedaf] with SbfI and NotI, blunting the cleaved termini with T4 DNA polymerase, and self-ligating the fragment containing the piggyBac transposon to produce pBac[3xP3-DsRedaf]SNKO, which contained a unique PstI site in the right piggyBac ITR.
The attB amplification product was cloned into pBac[3xP3-DsRedaf]SNKO at the PstI site in the piggyBac right ITR to produce pBattB[3xP3-DsRedaf]SNKO. The orientation of the attB sequence was verified by gene amplification to ensure that two functional sets of piggyBac ITRs would be produced upon integration into the attP sequence present at the An. stephensi 44C line docking site (Isaacs et al., 2012), which was generated using pBac[3xP3-ECFPfa]-attP (Fig. 1, Nimmo et al., 2006). The 3XP3-DsRed-SV40 expression cassette was restored by cloning the 3XP3-DsRed-SV40 expression cassette from pBac[3xP3-DsRedaf] into pBattB[3xP3-DsRedaf]SNKO through the unique BstBI and FseI sites in each construct to produce pBattB[3xP3-DsRedaf]. The nanos-driven piggyBac transposase expression cassette from pBac[3XP3-DsRed-SV40] 3′Nan-pBORF-3.8Nan5′ was cloned into pBattB[3xP3-DsRedaf] through the unique SbfI (in DsRed) and FseI (at the 5′-end of 3.8 nanos promoter) sites in each construct to produce the pBacDsRed-attB[3.8nanos-pBacORF] plasmid.
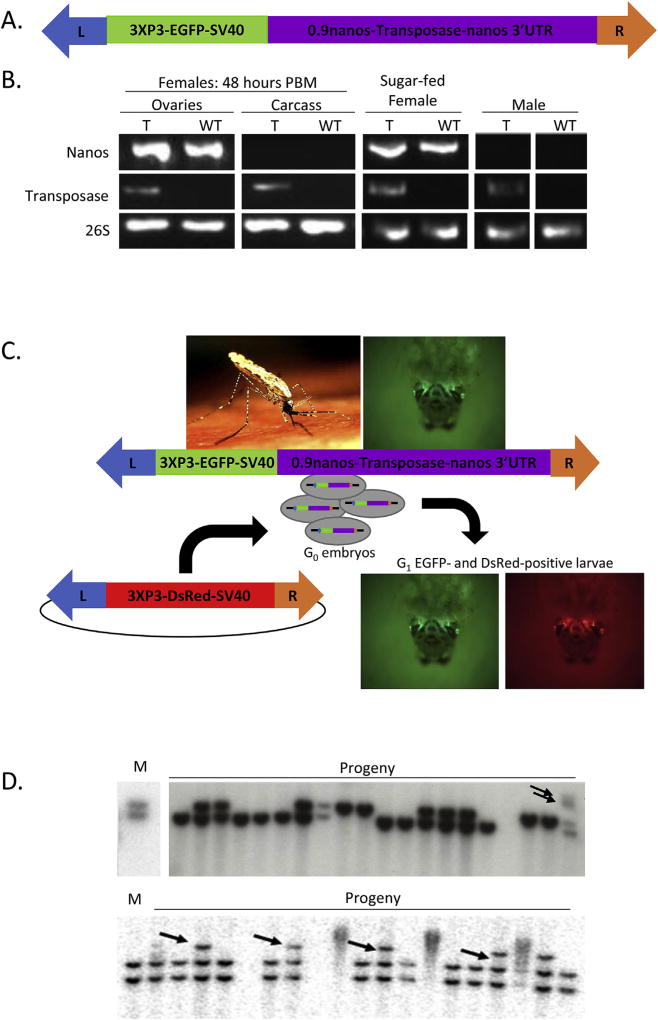
Fig. 1. Endogenously-encoded transposase-mediated remobilization.
A) Schematic representation of the piggyBac construct used to engineer a transgenic An. stephensi line expressing transposase driven by nanos control elements. The construct encodes EGFP with a 3XP3 promoter driving expression in the eyes and the Simian Virus 40 3′UTR, as well as the piggyBac transposase coupled to 0.9 kb of genomic DNA immediately 5′ of the nanos coding region and the nanos 3′-end UTR. Both open reading frames are encoded between the piggyBac Left and Right ITRs (L, R). B) RT-PCR analysis of the presence of nanos gene transcript, piggyBac transposase transcript and 26 S ribosomal protein gene as a positive control in female ovaries and carcass and males. WT: wild type; T: transgenic As28+. C) Schematic of assay for integration of a non-autonomous element. A plasmid (thin black line) encoding the 3XP3-DsRed-SV40 transgene between piggyBac left and right ITRs (L, R) was injected without an exogenous transposase source into As28 + embryos (gray ovals) laid by transgenic EGFP-positive females. These females (image upper left) contain an integrated transgene comprising piggyBac left and right ITRs (L,R) flanking a 3XP3-EGFP-SV40 marker gene adjacent to the piggyBac transposase open reading frame (Transposase) flanked by the nanos 0.9 kb promoter, and 5′ - and 3′-end genomic DNA. This strain has EGFP fluorescence visible in the larval eyes (image upper right). As28 + embryos contain transposase expressed from the transgenic construct and if this expression results in functional transposase in the germline, the DsRed construct will be integrated into the An. stephensi genome, resulting in EGFP and DsRed expression in the eyes of larvae (images on lower right). D) Southern blot analysis of two mothers (M) and their progeny using a 32P-labelled probe for EGFP. Diagnostic DNA fragments present in progeny but not mothers are indicated with arrows and represent remobilization events.
2.2. Mosquito transformation
Transgenic An. stephensi carrying pBac3XP3-EGFP[0.9nanos-pBacORF] were created by injecting pre-blastoderm embryos with a mixture of pBac3XP3-EGFP[0.9nanos-pBacORF] (300 ng/µL) and piggyBac helper (200 ng/µL) plasmids using procedures described in Nirmala et al. (2006). A transgenic An. stephensi line carrying pBac3XP3-DsRed[3.8nanos-pBacORF] was obtained using site-specific integration. Embryos from the docking line 44C (Amenya et al., 2010; Isaacs et al., 2012) were microinjected with a mixture of pBac 3XP3-DsRed[3.8nanos-pBacORF] (300 ng/µL) plasmid and φC31 integrase mRNA (400 ng/µL) as described (Nimmo et al., 2006).
2.3. Reverse transcriptase-PCR
Total RNA was isolated from whole animals and dissected tissues using RNAeasy Mini Kit (Qiagen) and treated with DNAse I (Invitrogen). The OneStep RT-PCR kit (Qiagen) was used for amplification of diagnostic products using primers listed in Supplemental Table 1. The reaction mixture was incubated at 50 °C for 30 min and 95 °C for 15 min. Amplification conditions were 94 °C for 1 min followed by 30 cycles of 94 °C for 1 min, 56 °C for 1 min and 72 °C for 1 min with a final extension for 10 min at 72 °C.
2.4. Southern blot analyses
Samples of genomic DNA from individual mosquitoes were isolated using Wizard Genomic DNA Purification kit (Promega) for Southern blot analyses and ~3.5 µg was digested using 30U of EcoRI or KpnI in a 20 µL reaction. Digested DNA was resolved in a 0.8% agarose gel in Tris borate EDTA buffer (TBE: 0.089M Tris, 0.089M borate, 2 mM EDTA) at 70 V for 5 h or at 20 V overnight (~16 h). Gels were visualized after a 10-min stain in a GelRed 3,000X (Biotium), Tris acetate EDTA (TAE) solution (0.04M Tris, 0.04M acetate, 1 mM EDTA), soaked in a denaturation solution (1.5M NaCl, 0.5 NaOH) twice for 15 min, and soaked in a neutralization solution (1.5M NaCl, 0.5M Tris-HCL (pH 7.2), 1 mM EDTA) twice for 15 min. Gels were rinsed with deionized water. DNA transfer to a nylon membrane was set up according to standard protocols (Sambrook et al., 1989). Following transfer, nylon membrane blots were rinsed in 2X saline sodium citrate (SSC) and cross-linked at 1200 )µW/cm2 in a UV Stratalinker (Stratagene). To generate probes to detect insertions of the gene encoding the enhanced green fluorescent protein (EGFP) in the As28 + line, the first 450bp of the EGFP ORF were cloned into a TOPO TA vector (Invitrogen) generating the TOPO-EGFP plasmid, which then was digested with EcoRI. The resulting fragment containing the EGFP ORF DNA sequence was extracted and purified from an agarose gel.
Southern blot analysis also was used to identify G17 males from the 3.8nanos-attP44c colony that had one copy of each of the genes encoding the enhanced cyan fluorescent protein (ECFP) and Discosoma species Red (DsRed) marked constructs. Genomic DNA isolated from individual males was divided such that one-half was digested with KpnI and probed with 32P-labelled ECFP DNA probe, and the remainder digested with EcoRI and probed with the 32P-labelled DsRed DNA probe. Labelled probes targeting ECFP or DsRed genes were generated by amplification of 600–700 bp fragments from plasmids bearing the marker genes (primers listed in Supplemental Table 1).
The amplification or digestion products were gel-electrophoresed, extracted and amplified with 32P-labelled dATP and dCTP (Perkin-Elmer) using Megaprime DNA labeling system (Amersham).
2.5. Splinkerette PCR
Splinkerette PCR was performed as described previously (Potter and Luo, 2010). Genomic DNA was extracted from adult mosquitoes using DNeasy Blood & Tissue (Qiagen) or Wizard Genomic DNA Purification kits (Promega) and digested with BstI. Amplification products were resolved in agarose gels; two fragments were amplified from the dual reporter construct at the 44C site with the piggyBac5Rev primer and were ~350 and ~400bp in length, respectively. Any fragment of any other size was considered diagnostic for remobilization. All fragments were gel-extracted, purified and sequenced. Diagnostic amplifications to identify the genotype of mosquitoes derived from the dual-reporter line with exceptional phenotypes were performed with primer combinations as depicted in Fig. 3 with primers listed in Supplemental Table 1.
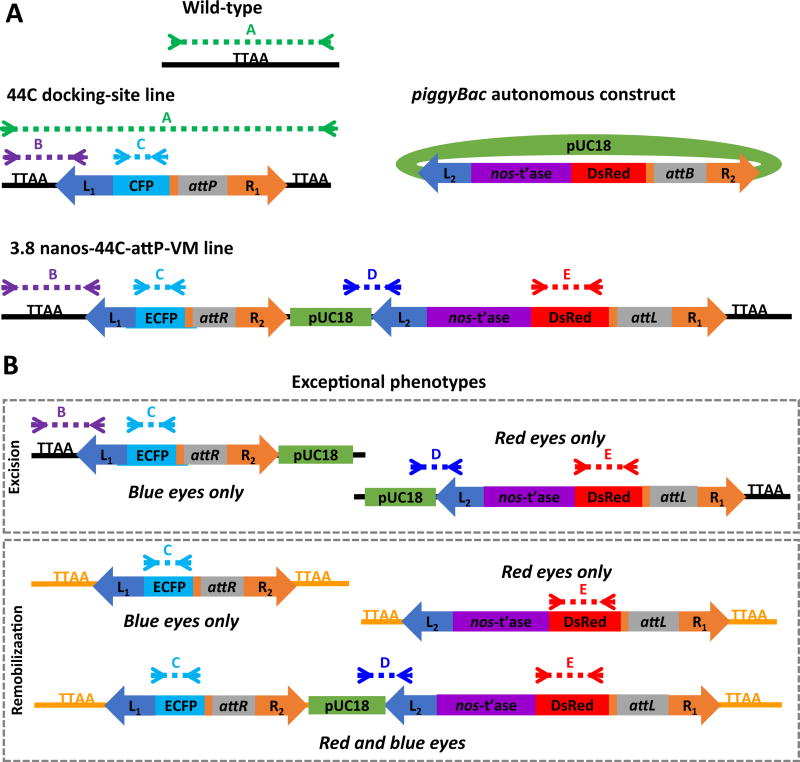
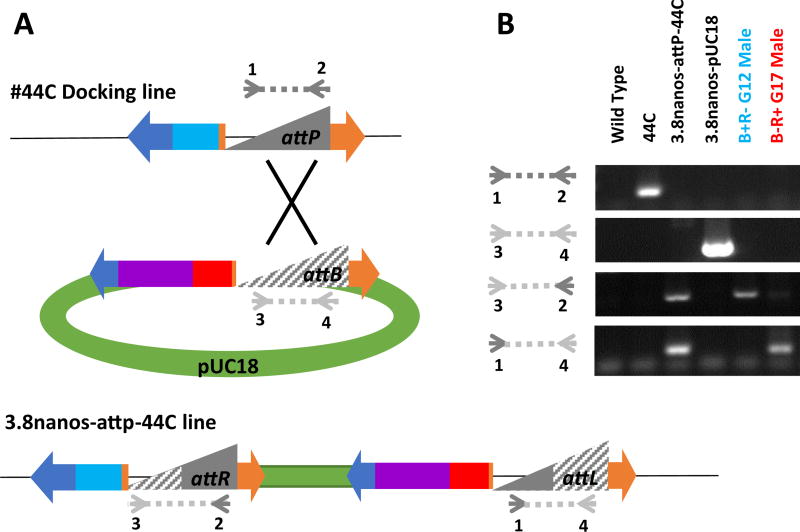
Fig. 3. Schematic representations of gene structures and expected amplification products resulting from autonomous mobility of transgenes.
A) The 44C docking site line comprises a transgene consisting of the left (L1) and right (R1) inverted terminal repeats of the piggyBac transposable element flanking the coding sequence of the enhanced cyan fluorescent protein (ECFP) driven by a 3XP3 promoter and sequences for site-specific recombination (attP) from the phage φC31 (Isaacs et al., 2012) integrated into the genome (horizontal black line). piggyBac transposase-mediated insertion causes a target-site duplication (TTAA) flanking the transgene. φC31 recombinase-mediated integration of a piggyBac autonomous construct comprising a cloning plasmid (pUC18) carrying an additional set of piggyBac left (L2) and right (R2) inverted terminal repeats, the piggyBac transposase coding sequence driven by the nanos promoter (nos-t’ase), the DsRed fluorescent protein coding sequence (DsRed) driven by a 3XP3 promoter and the φC31 attB (attB) site yields the 3.8 nanos-attP44C VM line. DNA derived from wild-type and 44C docking-site line mosquitoes generates diagnostic amplification fragments of 384 and 4396 bp, respectively, when using primer set A. Similarly, primer sets B and C yield fragments of 404 and 654 bp, respectively, when DNA samples from 44C docking-site and 3.8 nanos-attP44C mosquitoes are used as templates. Primer sets D and E are specific to the 3.8 nanos-attP44C line and produce fragments of 182 and 621 bp, respectively. B) The generation of exceptional phenotypes (Blue or Red eyes only) can result from excision or remobilization of portions of the 3.8 nanos-attP44C transgene complex. Primer sets B, C, D and E are diagnostic for excision events in the combinations illustrated. Remobilization puts portions of or the entire transgene complex at a new site in the genome (horizontal orange line). Inverse PCR techniques are used to identify the new genomic location.
2.6. Inverse PCR
Inverse PCR was performed as described previously (Handler et al., 1998) with primers listed in Supplemental Table 1. Inverse PCR and splinkerette PCR are analogous procedures that provide the same information, however we found that splinkerette PCR was consistently effective for identifying the DNA flanking the piggyBac left ITR, whereas Inverse PCR was more reliable for identifying the DNA flanking the piggyBac right ITR. The Inverse PCR protocol was performed initially to identify individual mosquitoes that had been observed to have only one fragment for each ECFP and DsRed genes after probing of the genomic DNA by Southern blot. Following screening for exceptional phenotypes, genomic DNA was extracted from individual mosquitoes and inverse PCR performed on pools of genomic DNA aliquots from individuals of the same phenotype. Digests were performed with either HeaIII or MspI for two hours and purified by ethanol precipitation. Ligations were performed using T4 DNA ligase (New England Biolabs) with a total reaction volume of 400 µL, overnight at 4 °C. DNA from ligation reactions was purified by ethanol precipitation and used as a template for gene amplification with primers listed in Supplemental Table 1. Amplification products were run on agarose gels and diagnostic fragments extracted using QiaQuik Gel Extraction kit (Qiagen), cloned into pSC-B-amp/kan using StrataClone Blunt PCR Cloning kit (Agilent Technologies) and transformed into Mix & Go Competent Escherichia coli strain JM109 (Zymo). Resulting bacterial colonies were picked and grown for plasmid amplification, and DNA isolated using Zyppy Plasmid Miniprep kit (Zymo) and sequenced by Laguna Scientific, a local fee-for-service company.
3. Results
3.1. A synthetic autonomous transposon; mobilization detected by Southern blot analyses
As a first step towards generating a synthetic autonomous construct, the piggyBac transposon was used as the basis for a transgene cassette that encoded EGFP as a marker gene and the piggyBac transposase under the control of the An. stephensi nanos promoter and 5′- and 3′ -end UTR (Fig. 1A). Work in the fruit fly, Drosophila melanogaster, showed that the nanos promoter and 5′ -end UTR drive expression of its mRNA in the maternal follicle cells and the 3′-end UTR mediates localization of the transcripts to the germline (Ali et al., 2010; Chen and McKearin, 2003; Curtis et al., 1995; Doren et al., 1998; Gavis et al., 1996; Gottlieb, 1992; Tracey et al., 2000). Orthologous mosquito genes appear to share similar expression characteristics (Calvo et al., 2005; Adelman et al., 2007; Meredith et al., 2013). Following transformation of An. stephensi, a line designated As28 + with four separate copies of the integrated construct was chosen for further analyses (Jimenez, 2009). RT-PCR analysis revealed that unlike the products of the endogenous nanos gene, the recombinant transposase was expressed in all tissues assayed including female somatic and male tissues (Fig. 1B). To assay this line for transposase activity, embryos were injected with an additional construct conferring DsRed expression in the eyes. The DsRed gene was flanked by the piggyBac left and right ITRs, but no transposase was encoded on the construct and no helper was introduced during the microinjection (Fig. 1C). Integration of this second construct was seen at a rate (4 insertions from 430 embryos injected, 0.93%) comparable to that observed with helper plasmid injection (17 insertions from 1000 embryo injections,1.7%) showing that the transposase expressed from the As28 + transgenic mosquitoes was active in the germline and so we expected that the construct would be able to self-mobilize.
A series of Southern blot analyses were performed on genomic DNA collected from individually outcrossed transgenic mothers and their offspring to assess whether any new insertions of the construct could be identified in progeny that were not present in the mother. Such insertions would be apparent by additional diagnostic DNA fragments or fragments of differing size in Southern blot analyses, and would be suggestive of transposase-mediated remobilization events. Two such events were detected in 386 progeny originating from 21 mothers (Fig. 1D). As each of the progeny samples results from a single gamete from each mother, we can calculate a remobilization frequency of 0.52% or 0.0052 remobilizations per gamete in this assay.
3.2. A dual-marker transgenic line, 3.8nanos-attP44C, for visual detection of remobilization
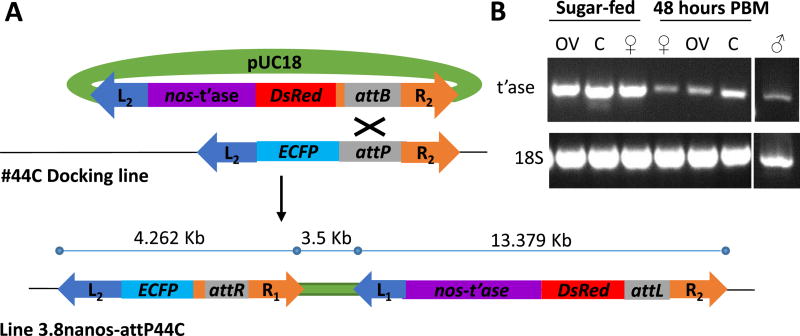
A new remobilization construct was generated with a number of improved characteristics. First, the final construct included two marker genes, ECFP and DsRed, each flanked by a set of piggyBac ITRs, such that the marker genes were linked tightly and could be remobilized either jointly or separately (Fig. 2A). Excision and remobilization events segregating the two fluorescent markers could be identified by visual screening of the eyes allowing the selection of individuals with relevant phenotypes (only one of the markers expressed in the eyes) for further molecular characterization. Second, the amount of 5′ flanking DNA from the putative nanos gene promoter was increased from 0.9 to 3.8 kb based on the previous observation that the former was not sufficient to specify tissue- and sex-specific expression characteristics. DNA fragments of ~1.5 kb comprising the putative promoter and 5′-end sequences of the Ae. aegypti and An. gambiae nanos orthologs were shown previously to be sufficient to drive abundant sex- and tissue-specific expression (Adelman et al., 2007; Meredith et al., 2013). However, following injection and recovery of transgenic mosquitoes, RT-PCR analysis of total RNA collected from sugar-fed and blood-fed whole females, dissected female tissues and whole males showed piggyBac transposase transcript present in all samples assayed (Fig. 2B). Third, a transformation scheme was devised to integrate a single copy of the gene-drive construct using φC31-mediated recombination to place the construct at a known and characterized location in the genome (Fig. 3). The transgenic line bearing the construct was generated by microinjection of the An. stephensi docking site line 44C (Amenya et al., 2010; Isaacs et al., 2012). G1 individuals with both DsRed- and ECFP-fluorescent eyes were scored as positive for integration of the construct into the 44C site. A line, 3.8nanos-attP44C, was derived from intercrossing these individuals and was maintained initially with screening at every generation.
Fig. 2. Generation and expression characterization of 3.8nanos-attP44C, a transgenic An. stephensi dual-reporter for transposon mobilization.
A) Schematic representation of the transgenic design. DsRed, the piggyBac transposase open reading flanked by 3.8 kb of the 5′ region of An. stephensi nanos and the nanos 3′untranslated region (nos-t’ase) and a φC31 attB site were encoded between piggyBac ITRs and the plasmid was used to transform attP44C line with φC31 integrase to generate two tightly-linked markers. B) RT-PCR analysis of transposase expression in 3.8nanos-attP44C in sugar-fed (never blood-fed), 48 h post-blood meal (PBM) females (OV: ovary; C: carcass) and males.
The fluorescent marker arrangement allowed us to separate individuals with evidence of putative movements during larval screening by identifying those with the exceptional phenotypes of only ECFP or only DsRed fluorescence in the eyes (Fig. 3). Genomic DNA samples from exceptional individuals were analyzed by gene amplification to verify the observed phenotype and to characterize remobilization events. This diagnostic gene amplification scheme allowed us to characterize individuals as excision or remobilization events since an exceptional phenotype could arise by excision of the other marker gene, or a remobilization of a marker gene to a new genomic location and segregation from an occupied ECFP-marked 44C docking site.
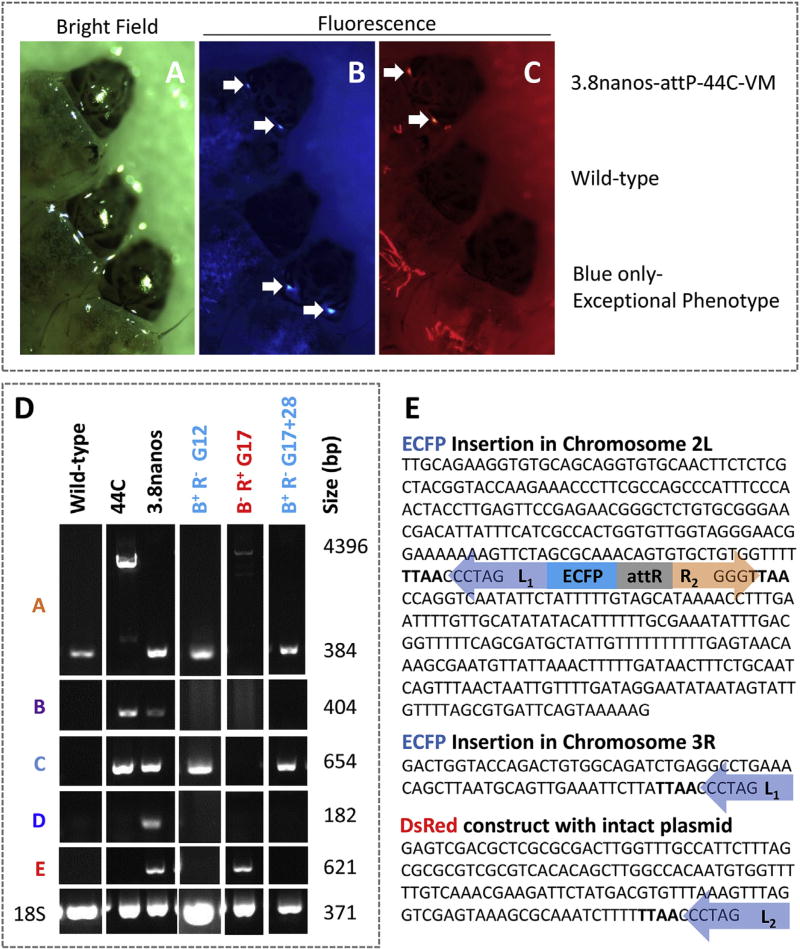
A number of individuals with exceptional phenotypes were identified immediately following line establishment and we suspected some of these to have resulted from expression of the transposase from the injected plasmid. φC31-mediated insertion at the 44C site was validated by gene amplification and DNA sequencing, confirming the desired dual-marker construct illustrated in Fig. 3. However, it is possible that the piggyBac transposase encoded in pBacDsRed-attB[3.8nanos-pBacORF] was expressed during the initial injection and it could have mediated additional insertions at other genomic locations. An ECFP-only individual was identified in generation 12 (G12) and analysis of its DNA by gene amplification profiling and splinkerette PCR showed that the ECFP construct was at a genomic location other than the 44C integration site (Fig. 4A – D). The integration was confirmed by sequencing to be on chromosome 3R at a TTAA site on scaffold location 1480193–14 80196 (Fig. 4E; Indian Strain, VectorBase.org). Additionally, a G17 DsRed-only individual was characterized by gene amplification and inverse PCR (analogous to splinkerette PCR) to be the result of an excision of the ECFP construct (Fig. 4D and E).
Fig. 4. ECFP construct mobilization.
Mosquitoes of the 3.8nanos-attP44C and -VM lines were screened for ECFP and DsRed in the eyes (white arrows). From top to bottom in each picture: 3.8nanos-attP44C-VM larvae with both fluorescent markers, a larva from the wild type colony, a 3.8nanos-attP44C-VM larvae with an exceptional phenotype (ECFP, but not DsRed). A) Bright field microscopy B) ECFP filter C) DsRed filter D) DNA derived from wild-type, 44C docking site line and 3.8 nanos-attp44cVM line mosquitoes, and exceptional individuals amplified with the designated primer sets (Fig. 3) produces amplicons supporting remobilization of the ECFP portion of the transgene complex. E) DNA sequence flanking the piggyBac left and right ITRs identified by inverse PCR in exceptional individuals. Abbreviations: attB, attL, attP and attR, φC31 attachment sites bacteria, left, phage and right, respectively; ECFP, enhanced cyan fluorescent protein; DsRed, Discosoma sp. red fluorescent protein; L1,2, R1,2, left and right piggyBac inverted terminal repeats, respectively; nos-t’ase, nanos promoter and piggyBac transposase open-reading frame; TTAA, piggyBac transposon recognition tetranucleotide.
To confirm that the ECFP integration on chromosome 3R was mediated by transposition out of the 44C docking site following its prior φC31-mediated integration and not an insertion mediated by transposase expressed during injection of G0 embryos, a set of gene amplification primers were designed to differentiate the attR site that would be present on the ECFP construct in the former from the attB site that would be present in the latter (Fig. 5A). Gene amplification analysis identified an attR site and not an attP site in the DNA of the ECFP-only individual, confirming this event as a remobilization event (Fig. 5B). Similarly, gene amplification showed the presence of an attL site in the genomic DNA of the G17 DsRed-only individual leaving the DsRed construct behind, indicating that the ECFP portion of the construct was mobilized out of the site (Fig. 5B). We can conclude from these data that remobilization occurred at least once by generation 17.
Fig. 5. Verification of mobilization out of the 44C genomic location.
A) Gene amplification scheme to differentiate the presence of a φC31 attP site present in the docking line and attB site present on the injected plasmid from attR and attL sites. Gene-specific primers are shown as arrowheads flanking a diagnostic fragments represented as a dotted line. B) PCR analysis on individuals with exceptional phenotypes, ECFP- or DsRed-only, collected at generation 12 (G12) and generation 17 (G17) respectively. Wild-type An. stephensi were used as a negative control and ECFP-only mosquitoes from docking line 44C and ECFP/DsRed mosquitoes from line 3.8nanos attP44C were used as positive controls for attP and attR sites respectively. The injected plasmid pBacDsRed-attB[3.8nanos-pBacORF] was used as a positive control for the attB site.
3.3. Identification and characterization of remobilization in 3.8nanos-attP44C-VM
In order to ensure that individuals identified as only ECFP-positive or only DsRed-positive in future generations represented transposase activity originating from expression of the transgene located at the 44C site, it was necessary to isolate the φC31 integrase-mediated site-specific integration from genotypes resulting from G0 transposase-mediated remobilization and subsequent mobilizations being maintained in the colonies. Eight G17 males from 3.8nanos-attP44C were outcrossed individually and then assayed molecularly for their genotype. Male #3 was found to have only one copy of ECFP and DsRed by Southern blot analysis and inverse PCR showed that they were both present at the 44C site as originally intended (Supplemental Fig. 1). Progeny of male #3 were used to found the line 3.8nanos-attP44C-VM. Generation number will be counted as a continuation of line 3.8nanos-attP44C, so that the founding generation of 3.8nanos-attP44C-VM is G17.
To identify remobilization events, 3.8nanos-attP44C-VM males were outcrossed at every generation from G17 for five generations and the progeny screened for exceptional phenotypes, which include not only mosquitoes showing only ECFP or DsRed, but also ECFP- and DsRed-positive male individuals. The 44C site is located on the X-chromosome, so by outcrossing males, we can identify any male progeny with fluorescence as exceptional, since the original insertion should only be passed to females. After five generations of outcrossing, no exceptional phenotypes were seen. We reasoned that the mobilization frequency could be influenced by transposase dose, so we began maintaining the line by intercrossing. Larvae were screened at every generation for five additional generations without identification of exceptional phenotypes.
After 28 generations, four G45 males from the transgenic line were outcrossed and 38 out of 449 total progeny were ECFP-only individuals (8.4%, Fig. 4A, B and C). In order to identify how many remobilization events had occurred over the past generations 10 ECFP-only males were outcrossed to generate individual families to increase the amount of genomic DNA available for analysis.
Only one remobilization event was captured; a remobilization of the ECFP-marked portion of the construct was identified by inverse PCR to be on chromosome 2 L, scaffold_00100 (Indian Strain, VectorBase.org, Fig. 4E) at the TTAA sequence at position 272403–272406. TTAA sequences flanking the construct support the conclusion that the construct was precisely excised and reintegrated by piggyBac transposase (Fraser et al., 1996).
4. Discussion
We report here on the activity in An. stephensi of transposon-based synthetic autonomous gene drive constructs, As28 + and 3.8nanos-attP44C, encoding piggyBac transposase under the control of the An. stephensi nanos gene promoter, 5′- and 3′-end UTRs. The recovery of these constructs at genomic loci other than those of their original insertion sites is a proof-of-principle that a synthetic transposon construct can be designed to self-mobilize. We observed two mobilization events in the As28 + line in 21 mother-progeny sets and this represents an estimated remobilization frequency of 0.0052/gamete. In the 3.8nanos-attP44C lines, we detected two events over 45 generations at what must be a much lower frequency per gamete. Thus, it appears that remobilization occurred at a higher rate in the As28 + line than in the 3.8nanos-attP44C lines. An alternative mechanism of relocation by recombination was ruled out in the exceptional individuals recovered from 3.8nanos-attP44C because target site (TTAA) duplications were detected flanking the right and left ITRs of the remobilized construct, supporting the conclusion that mobilization occurred by a cut-and-paste mechanism into the TTAA target site typical for piggyBac insertions (Fraser et al., 1996). We did not observe mobilization of the DsRed-marked construct in the 3.8nanos-attP44C transgenic lines. While the overall number of mobilizations observed is too small to be conclusive, it may be that this construct was prohibitively large. However, the piggyBac transposase has been demonstrated in other systems to faithfully remobilize a lengthy cargo, up to 100 kb (Li et al., 2011; Ding et al., 2005). It will be important to demonstrate that a transposon-based gene drive will be able to accommodate cargo in mosquitoes in order to drive anti-pathogen effector molecules into target populations.
The difference in mobilization frequencies between the two transgenic lines is puzzling and likely reflects a contribution of several possible factors. Perhaps some events were not captured in the 3.8nanos-attP44C lines due to mosquito line maintenance requirements. For example, a rare event may not persist in the colony for more than one generation since only a proportion of total mosquito embryos produced each generation are used to maintain the colony. In the As28 + line, mother progeny sets were analyzed, so detection did not necessitate the mobilization being frequent in the colony.
Going forward with the development of a usable piggyBac-based gene-drive in An. stephensi, it will be important to identify the contribution of transposase dose and genomic context to construct mobility. The As28 + transgenic line contained four transposase gene copies, whereas the 3.8nanos-attP44C lines had only one, and increased transposase abundance could explain the higher frequency of construct self-mobilization. However, the piggyBac element does not mobilize in Ae aegypti, even in the presence of functional transposase (Palavesam et al., 2013; Sethuraman et al., 2007). Furthermore, piggyBac element mobilization in D. melanogaster has been attributed to genomic context, not transposase abundance, while no genomic context has been identified that supports remobilization in Ae. aegypti (Esnault et al., 2011; Palavesam et al., 2013; Sethuraman et al., 2007).
The assay in which a plasmid encoding a DsRed marker gene between the piggyBac left and right ITRs was injected without a transposase source into the As28 + transgenic line resulted in efficient integration of the marked construct, showing that functional transposase is expressed from the integrated transgene and is capable of mediating integration at a frequency comparable to that of the injected helper plasmid in the original line generation. This integration frequency (0.97%) and the remobilization frequency observed in this line (0.52%) also are within the same order of magnitude. If integration of a piggyBac construct using a similar assay in the 3.8nanos-attP44C line is higher than the observed remobilization rate, we would suspect either transposon regulation at the level of genome integration or a difference in the make-up of the construct instead of a difference due to transposase dose. This could be explored by insertion of the constructs into alternative genomic locations that may support a more robust expression of the transposase; a number of An. stephensi lines bearing attP sites at different locations are available that could be used to test this (Amenya et al., 2010; N. Jasinkiene, personal communication).
Heritable piggyBac construct mobilization may be influenced by the amount of available transposase in the germline tissue. We observed that, unlike the Ae. aegypti and An. gambiae nanos control elements (Adelman et al., 2007; Meredith et al., 2013), the nucleotides up to nearly 4 kb to the 5′-end of the transcription start site of An. stephensi nanos were not sufficient to drive tissue-specific expression of the transposase, but induced expression in both germline and somatic female tissue and in males. A more restricted promoter that drives accumulation of piggyBac transposase to a high level in the ovaries may increase germline transposition frequency.
Synthetic, non-autonomous piggyBac constructs can be mobilized in the An. stephensi genome using a ‘jumpstarter’ helper line expressing the piggyBac transpose (O’Brochta et al., 2011). This enhancer-trap mechanism supported the hypothesis that a piggy-Bac transposase expressed from the An. stephensi genome could remobilize synthetic constructs in the genome to a new location, and a synthetic construct comprising a piggyBac transposase gene cloned between the piggyBac left and right ITRs in theory could remobilize itself. Our data validate that this is possible. However, remobilization frequencies in all lines are far below the 10% transposition per insert per generation that is predicted by models to be necessary for a useful gene-drive mechanism (Ribeiro and Kidwell, 1994; Rasgon and Gould, 2005; Marshall, 2008).
Crossing our lines with a helper line reported by O’Brochta et al. (2011) may provide additional insight into why mobility in our line is so low. Furthermore, while transposase transcript and protein were not quantified in the helper lines, such analyses may give an insight into the contribution of transposase dose to construct mobility.
It is also possible that the transposase is being post-transcriptionally regulated by the Piwi-interacting RNA (piRNA) pathway. piRNAs are derived from regions of the genome that have remnants of transposons to which the species was exposed previously in its evolutionary history. It may be that an ancestor of our An. stephensi line encountered one or more members of the piggyBac transposon family. A BLAST of the currently available assembled and annotated An. stephensi genomes identifies regions 19 to 40 nucleotides in length with 86–100% similarity to piggyBac transposase (VectorBase.org), but data are not currently available as to whether these regions are piRNA producers. These results are consistent with data from other anopheline species that contain regions in their genomes with similarity to piggyBac transposase (Fernandez-Medina et al., 2011; Marinotti et al., 2013). It could be that ancient exposures to ancestral piggyBac transposons provide enough sequence to establish repression of the contemporary transposon. If indeed piRNA regulation was influencing transposase availability, the construct can be redesigned to avoid this regulation by altering the nucleotide sequence. Since the regulation is mediated by nucleotide sequence specificity, if the transposase is encoded with different codons, the transposase transcript would essentially be invisible to the piRNA machinery.
In addition to modifications to improve tissue specificity, optimize transposon dose and genomic context and to avoid post-transcriptional regulation, alterations to the encoded transposase itself may improve construct mobility. A hyperactive piggyBac transposase has been reported to increase transposition nine-fold in mammalian cells (Yusa et al., 2011); it would be interesting to see whether expression of this transposase from our construct would yield a more efficient transposase-based genetic drive.
The piggyBac element has been the most successful transposon-based platform so far for genetic applications in Anopheles species, but it is possible that another transposable element would be more useful. Transposons have been sought from other organisms that have mobility in Anopheles species and Ae. aegypti with the hope of applying them as tools for genetic manipulation, including gene-drive and molecular genetic studies. However, specific transposable elements do not exhibit the same mobility characteristics in mosquitoes as they do in D. melanogaster and other insect species (O’Brochta et al., 2003; Wilson et al., 2003; Palavesam et al., 2013; Scali et al, 2007; Sethuraman et al, 2007). For example, P elements can mediate transposition of a transgene into the D. melanogaster genome, but are not effective for transformation of any mosquito species. The Minos element, which is remobilized following integration into the D. melanogaster genome in both somatic and germline tissues, moves only in the soma in An. stephensi (Scali et al, 2007). A Herves element isolated from An. gambiae, and later identified in wild populations of this species as well as An. merus and An. arabiensis, appears to be active in An. gambiae (Arensburger et al, 2005). An autonomous construct based on it could be tested for mobility in An. stephensi and An. gambiae.
It is possible that inundative releases of transgenic mosquitoes may be sufficient for the goal of fixation of an anti-pathogen genotype into wild populations, but the predicted benefits afforded by gene-drive technology in terms of reduced cost and effort of implementation over an inundative release strategy, makes developing the technology a worthwhile pursuit (Macias and James, 2015). Synthetic gene-drive systems based on homing endonucleases, MEDEA and on CRISPR/Cas9 biology have been demonstrated (Windbichler et al, 2011; Akbari et al, 2013; Gantz et al, 2015; Hammond et al, 2015). A Cas9-based system in An. stephensi with a homology-directed repair conversion frequency of ≥0.97 per gamete (Gantz et al, 2015) was at least two orders of magnitude more efficient than what we observed here. The feasibility of using a gene-drive technology for population level impact depends on developing and testing new genetic technologies in mosquitoes, and is supported by a diverse set of techniques for genetic manipulation. The successful design of an active, albeit low frequency, transposon-based synthetic autonomous gene-drive element, affords an additional option to approach population modification.
Supplementary Material
Acknowledgments
We thank Judy Coleman for help in maintaining the As28+, 3.8nanos-attP44C and 3.8nanos-attP44C-VM mosquito lines. Funding: this work was supported by a grant from the NIH NIAID (AI29746).
Abbreviations
- ECFP
Enhanced cyan fluorescent protein
- EGFP
Enhanced green fluorescent protein
- DsRed
Discosoma sp. red fluorescent protein
- Gx
Post-injection generation number x
- pBac
piggyBac
Footnotes
Appendix A. Supplementary data
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.ibmb.2017.06.014.
References
- Adelman ZN, et al. nanos gene control DNA mediates developmentally regulated transposition in the yellow fever mosquito Aedes aegypti. Proc. Natl. Acad. Sci. U. S. A. 2007;104(24):9970–9975. doi: 10.1073/pnas.0701515104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbari OS, et al. A Synthetic Gene Drive System for Local, Reversible Modification and Suppression of Insect Populations. 2013 doi: 10.1016/j.cub.2013.02.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali I, et al. Cis-regulatory elements affecting the Nanos gene promoter in the germline stem cells. J. Biotechnol. 2010;145(4):323–329. doi: 10.1016/j.jbiotec.2009.12.011. [DOI] [PubMed] [Google Scholar]
- Amenya DA, et al. Comparative fitness assessment of Anopheles stephensi transgenic lines receptive to site-specific integration. Insect Mol. Biol. 2010;19(2):263–269. doi: 10.1111/j.1365-2583.2009.00986.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arensburger P, et al. An active transposable element, Herves, from the African malaria mosquito Anopheles gambiae. Genetics. 2005;169(2):697–708. doi: 10.1534/genetics.104.036145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo E, et al. Nanos (nos) genes of the vector mosquitoes, Anopheles gambiae, Anopheles stephensi and Aedes aegypti. Insect Biochem. Mol. Biol. 2005;35(7):789–798. doi: 10.1016/j.ibmb.2005.02.007. [DOI] [PubMed] [Google Scholar]
- Chen D, McKearin DM. A discrete transcriptional silencer in the bam gene determines asymmetric division of the Drosophila germline stem cell. Development. 2003;130(6) doi: 10.1242/dev.00325. [DOI] [PubMed] [Google Scholar]
- Curtis D, Apfeld J, Lehmann R. Nanos is an evolutionarily conserved organizer of anterior-posterior polarity. Development. 1995;121(6) doi: 10.1242/dev.121.6.1899. [DOI] [PubMed] [Google Scholar]
- Ding S, et al. Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell. 2005;122(3):473–483. doi: 10.1016/j.cell.2005.07.013. [DOI] [PubMed] [Google Scholar]
- Doren M, Van Williamson AL, Lehmann R. Regulation of Zygotic Gene Expression in Drosophila Primordial Germ Cells. 1998 doi: 10.1016/s0960-9822(98)70091-0. [DOI] [PubMed] [Google Scholar]
- Esnault C, et al. Intrinsic characteristics of neighboring DNA modulate transposable element activity in Drosophila melanogaster. Genetics. 2011;187:319–331. doi: 10.1534/genetics.110.122168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Medina RD, Struchiner CJ, Ribeiro JM. Novel transposable elements from Anopheles gambiae. BMC Genomics. 2011;12(1):260. doi: 10.1186/1471-2164-12-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser MJ, et al. Precise excision of TTAA-specific lepidopteran transposons piggyBac (IFP2) and tagalong (TFP3) from the baculovirus genome in cell lines from two species of Lepidoptera. Insect Mol. Biol. 1996;5(2):141–151. doi: 10.1111/j.1365-2583.1996.tb00048.x. [DOI] [PubMed] [Google Scholar]
- Gantz VM, et al. Highly efficient Cas9-mediated gene drive for population modification of the malaria vector mosquito Anopheles stephensi. Proc Natl. Acad. Sci. 2015;112(49):E6736–E6743. doi: 10.1073/pnas.1521077112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavis ER, Curtis D, Lehmann R. Identification of cis-acting sequences that control nanosRNA localization. Dev. Biol. 1996;176(1):36–50. doi: 10.1006/dbio.1996.9996. [DOI] [PubMed] [Google Scholar]
- Gottlieb E. The 3’ untranslated region of localized maternal messages contains a conserved motif involved in mRNA localization. Proc. Natl. Acad. Sci. 1992;89:7164–7168. doi: 10.1073/pnas.89.15.7164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond A, et al. A CRISPR-Cas9 gene drive system targeting female reproduction in the malaria mosquito vector Anopheles gambiae. Nat. Biotechnol. 2015;34:78–83. doi: 10.1038/nbt.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handler AM, et al. The lepidopteran transposon vector, piggyBac, mediates germ-line transformation in the Mediterranean fruit fly. Proc. Natl. Acad. Sci. U. S. A. 1998;95(13):7520–7525. doi: 10.1073/pnas.95.13.7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn C, Wimmer EA. A versatile vector set for animal transgenesis. Dev. Genes Evol. 2000;210(12):630–637. doi: 10.1007/s004270000110. [DOI] [PubMed] [Google Scholar]
- Isaacs AT, et al. Transgenic Anopheles stephensi coexpressing single-chain antibodies resist Plasmodium falciparum development. Proc. Natl. Acad. Sci. U. S. A. 2012;109(28):E1922–E1930. doi: 10.1073/pnas.1207738109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James AA. Gene drive systems in mosquitoes: rules of the road. Trends Parasitol. 2005;21(2):64–67. doi: 10.1016/j.pt.2004.11.004. [DOI] [PubMed] [Google Scholar]
- Jimenez AJ. Characterization of Candidate Genes for Applications in Genetic Control Strategies of Mosquito-borne Diseases. University of California; Irvine: 2009. [Google Scholar]
- Kidwell M, Ribeiro J. Can transposable elements be used to drive disease refractoriness genes into vector populations? Parasitol. Today. 1992;8:325–329. doi: 10.1016/0169-4758(92)90065-a. [DOI] [PubMed] [Google Scholar]
- Labbé GMC, Nimmo DD, Alphey L. piggybac- and PhiC31-mediated genetic transformation of the Asian tiger mosquito, Aedes albopictus (Skuse) PLoS Neglected Trop. Dis. 2010;4(8):e788. doi: 10.1371/journal.pntd.0000788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MA, et al. Mobilization of giant piggyBac transposons in the mouse genome. Nucleic Acids Res. 2011;39(22):e148–e148. doi: 10.1093/nar/gkr764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macias VM, James AA. Impact of genetic modification of vector populations on the malaria eradication agenda. In: Adelman Z, editor. Genetic Control of Malaria and Dengue. Elsevier; 2015. pp. 423–444. [Google Scholar]
- Marinotti O, et al. The genome of Anopheles darlingi, the main neotropical malaria vector. Nucleic Acids Res. 2013;41(15):7387–7400. doi: 10.1093/nar/gkt484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall JM. A branching process for the early spread of a transposable element in a diploid population. J. Math. Biol. 2008;57(6):811–840. doi: 10.1007/s00285-008-0190-2. [DOI] [PubMed] [Google Scholar]
- Meredith JM, et al. Next-generation site-directed transgenesis in the malaria vector mosquito Anopheles gambiae: self-docking strains expressing germline-specific phiC31 integrase. PLoS One. 2013;8(3):e59264. doi: 10.1371/journal.pone.0059264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmo DD, et al. High efficiency site-specific genetic engineering of the mosquito genome. Insect Mol. Biol. 2006;15(2):129–136. doi: 10.1111/j.1365-2583.2006.00615.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nirmala X, et al. Functional characterization of the promoter of the vitel-logenin gene, AsVg1, of the malaria vector, Anopheles stephensi. Insect Biochem. Mol. Biol. 2006;36(9):694–700. doi: 10.1016/j.ibmb.2006.05.011. [DOI] [PubMed] [Google Scholar]
- O’Brochta DA, et al. Gene vector and transposable element behavior in mosquitoes. J. Exp. Biol. 2003;206(21) doi: 10.1242/jeb.00638. [DOI] [PubMed] [Google Scholar]
- O’Brochta DA, et al. piggyBac transposon remobilization and enhancer detection in Anopheles mosquitoes. Proc. Natl. Acad. Sci. U. S. A. 2011;108(39):16339–16344. doi: 10.1073/pnas.1110628108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palavesam A, et al. Post-integration silencing of piggyBac transposable elements in Aedes aegypti. In: Hansen IA, editor. PLoS One. 7. Vol. 8. 2013. p. e68454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter CJ, Luo L. Splinkerette PCR for mapping transposable elements in Drosophila. PLoS One. 2010;5(4):e10168. doi: 10.1371/journal.pone.0010168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasgon JL, Gould F. Transposable element insertion location bias and the dynamics of gene drive in mosquito populations. Insect Mol. Biol. 2005;14(5):493–500. doi: 10.1111/j.1365-2583.2005.00580.x. [DOI] [PubMed] [Google Scholar]
- Ribeiro JM, Kidwell MG. Transposable elements as population drive mechanisms: specification of critical parameter values. J. Med. Entomol. 1994;31(1):10–16. doi: 10.1093/jmedent/31.1.10. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: a Laboratory Manual, Second. Cold Spring Harbor Laboratory Press; New York: 1989. [Google Scholar]
- Scali C, et al. Post-integration behavior of a Minos transposon in the malaria mosquito Anopheles stephensi. Mol. Genet. Genomics. 2007;278(5):575–584. doi: 10.1007/s00438-007-0274-5. [DOI] [PubMed] [Google Scholar]
- Sethuraman N, et al. Post-integration stability of piggyBac in Aedes aegypti. Insect Biochem. Mol. Biol. 2007;37(9):941–951. doi: 10.1016/j.ibmb.2007.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tracey WD, et al. Quantitative analysis of gene function in the Drosophila embryo. Genetics. 2000;154(1) doi: 10.1093/genetics/154.1.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson R, et al. Post-integration behavior of a Mos1 mariner gene vector in Aedes aegypti. Insect Biochem. Mol. Biol. 2003;33(9):853–863. doi: 10.1016/s0965-1748(03)00044-4. [DOI] [PubMed] [Google Scholar]
- Windbichler N, et al. A synthetic homing endonuclease-based gene drive system in the human malaria mosquito. Nature. 2011;473(7346):212–215. doi: 10.1038/nature09937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusa K, et al. A hyperactive piggyBac transposase for mammalian applications. Proc. Natl. Acad. Sci. U. S. A. 2011;108(4):1531–1536. doi: 10.1073/pnas.1008322108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.