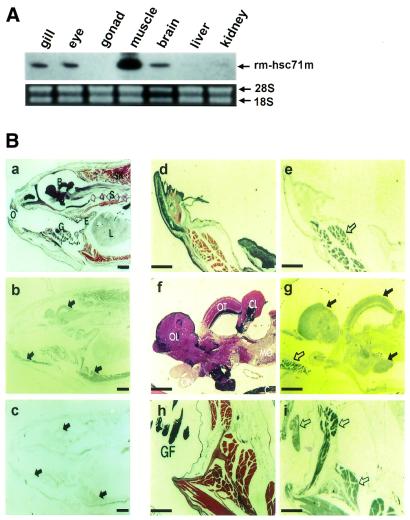
Figure 3.
rm-hsc71m expression is differentially regulated in a tissue-specific manner. (A) Expression profile of rm-hsc71m in adult tissues. Total RNAs were isolated from several organs of a 1-year-old fish and subjected to RT–PCR analysis using a set of rm-hsc71m-specific primers as described in the legend to Figure 2A. The lower panel shows photographs of ethidium bromide-stained 28S and 18S rRNAs as a loading control. (B) Views of paraffin-sectioned 1-year-old fish showing spatial expression patterns of rm-hsc71m detected by in situ hybridization with antisense RNA probes as described in Figure 2B. Sections were stained with hematoxylin/eosin (a, d, f and h) or hybridized with an antisense (b, e, g and i) rm-hsc71m RNA probe. As a negative hybridization control, a sense rm-hsc71m RNA probe was also employed (c). Arrows indicate regions showing strong expression of rm-hsc71m (open arrows in panels e, g and i indicate skeletal muscle tissue showing the strongest expression). B, brain; CL, cerebellum (corpus cerebelli); E, esophagus; GF, gill filament; I, infundibulum; L, liver; MO, medulla oblongata; O, oral cavity; OL, olfactory lobe (telencephalon); ON, optic nerve; OT, optic tectum; S, spine; SK, skeletal muscle. Scale bars: a–c, 1.25 µm; d–i, 0.32 µm.