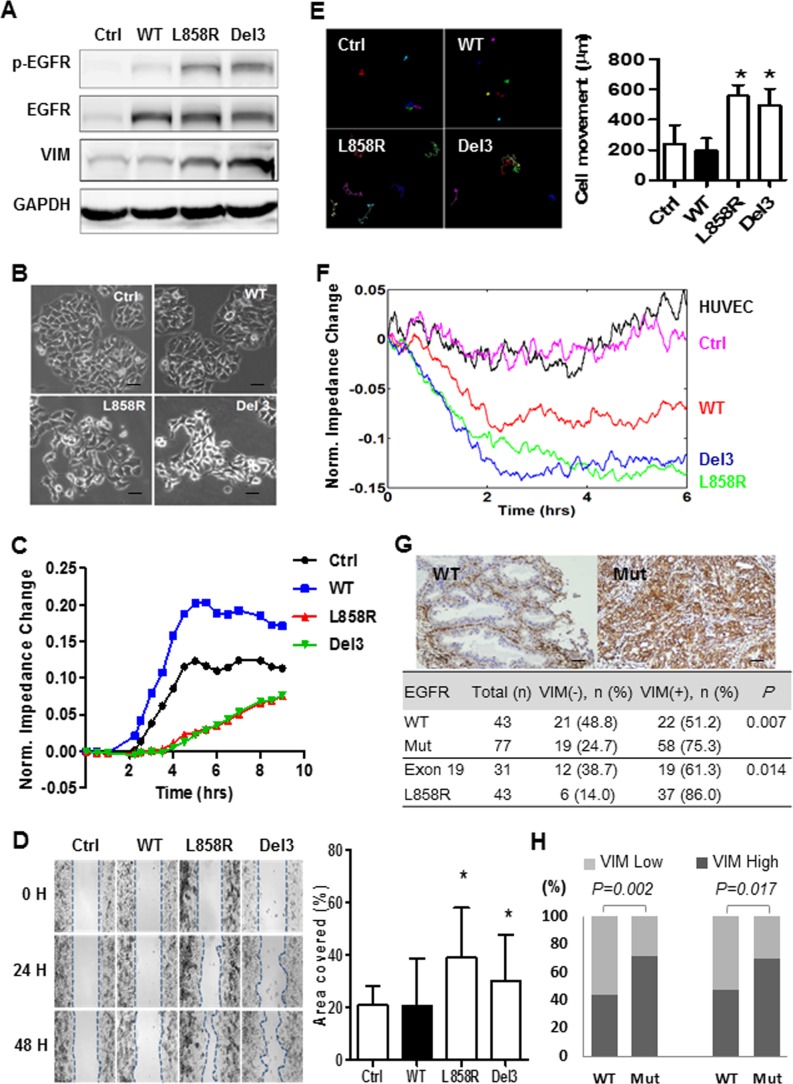
Figure 3. The mutated-EGFRs, exon 19 deletion and L858R point mutation, induce a mesenchymal-like phenotype and encourage cancer cell dissemination.
(A) Western blot analysis of phosphorylated EGFR (p-EGFR), total EGFR (EGFR), vimentin (VIM) and GAPDH in H1437 cells infected with lentiviral vectors encoding wild-type (WT) and mutated-EGFRs (L858R, a point mutation in exon 21; Del 3, an in-frame deletion in exon 19 of EGFR tyrosine kinase domain) or an empty control vector (Ctrl). (B) Representative phase-contrast images of H1437 cells infected with lentiviral vectors encoding WT and mutated-EGFRs (L858R or Del 3) or empty control vector (magnification = 100X, scale bar = 100 μm). (C) Electric cell-substrate impedance sensing (ECIS) analysis to monitor the changes in cell impedance as a result of the EGFR functional responses in H1437 cells infected with lentiviral vectors encoding WT and mutated-EGFRs or an empty control vector. (D) Representative images of the wound healing assay for H1437 cells infected with lentiviral vectors encoding wild-type (WT) and mutated-EGFRs (L858R or Del 3) or the empty control (Ctrl) vector (upper). The dotted lines indicate the wound edge at 0, 24 and 48 hrs, respectively (magnification = 50X). Quantitative analysis for the cells migrated during 48 hours (lower) (student t-test). * indicated significant at P < 0.05. (E) Representative images of cell tracking assay for H1437 cells infected with lentiviral vectors encoding WT and mutated-EGFRs or an empty control vector (upper). The colored lines represented the individual tracks of the motile cells (magnification = 100X, scale bar = 100 μm). Quantitative analysis for cell tracking assay (lower) (student t-test). * indicated significant at P < 0.05, ** indicated significant at P < 0.01. (F) An ECIS assay to monitor the changes in cell impedance in HUVEC cells as a result of the invasion of H1437 cells infected with lentiviral vectors encoding WT and mutated-EGFRs or an empty control (Ctrl) vector. (G) Association between vimentin (VIM) expression and EGFR mutation status in patients with lung adenocarcinoma. Representative pictures of the immunohistochemical analysis of VIM expression in adenocarcinoma specimens. A VIM-negative, WT EGFR adenocarcinoma case (upper left) and a VIM-positive, mutated-EGFR (Mut) adenocarcinoma case (upper right) are shown. The pictures were scanned at x 40 magnification (scale bar = 100 μm) and the size was adjusted to the screen. The distribution of VIT expression in the lung cancer specimens from the mutated- and WT-EGFR groups was compared using Fisher's exact test (bottom). (H) Correlation analysis of VIM expression with EGFR mutation status in mutated- and WT EGFR lung adenoacrinomas (Chi-square test). The sources of VIM expression levels of lung cancer and the corresponding clinicopathological parameters were downloaded from Chitale et.al. Memorial Sloan-Kettering Cancer Center (http://cbio.mskcc.org/public/lung_array_data/) (Left) and TCGA (https://tcga-data.nci.nih.gov/docs/publications/luad_2014/) (Right).