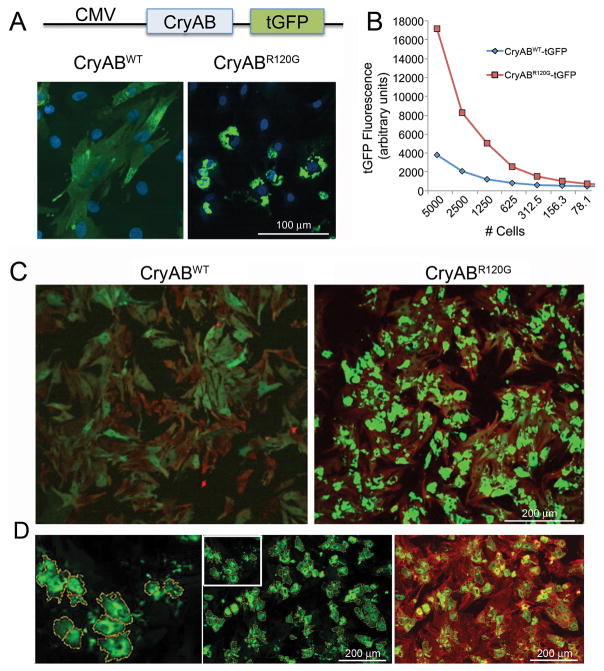
Figure 2. Aggregate induction and validation of the signal:noise ratio for CryABR120G-driven aggregate formation in primary cardiomyocytes.
A, High levels of either normal or mutant CryAB expression were driven off of the cytomegalovirus promoter after placement of the indicated construct into adenovirus. B, Quantitation of the relative signal strengths resulting from either CryABWT or CryABR120G was determined using immunofluorescence detected as a result of tGFP expression. C, Primary cardiomyocytes were plated at optimum densities for signal:noise generation, infected with either normal or mutant CryAB and the cells fixed and stained 3–4 days after infection. Cardiomyocytes were identified by immunofluorescent detection of troponin I (red) showing that >90% of the cardiomyocytes contained easily detectable (> 4-fold cytoplasmic CryABWTtGFP signal) CryAB-positive aggregates. D, Using BioTek Cytation 3 software, the algorithm defined aggregated protein on the basis of predefined filters and encircled them, creating yellow masks. To ensure measured aggregates were within cardiomyocytes as opposed to cardiac fibroblasts or endothelial contaminants, hierarchical detection was done using a cardiomyocyte-specific counterstain, TnI (red), such that only those cells where green aggregates were associated with red signal (green-over-red hierarchy) were scored and measured.