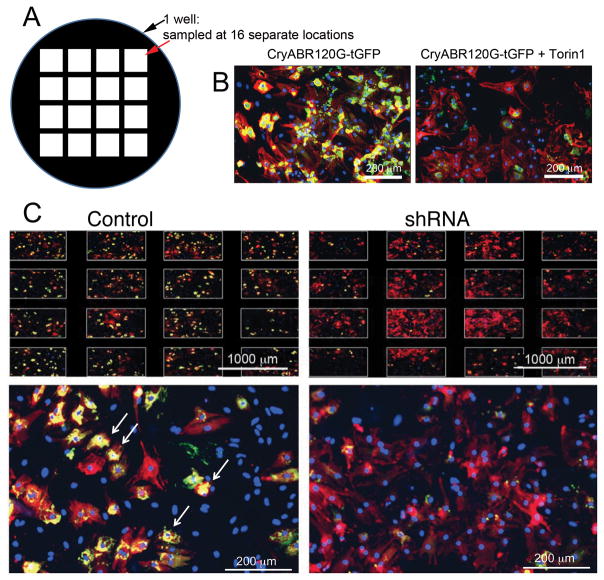
Figure 3. Inhibitory RNA knockdown of specific genes leads to decreased aggregate load in cardiomyocytes.
A, Each well in each multi-welled plate was sampled in 16 separate locations and only those plates showing uniform cardiomyocyte density and aggregates were selected for further analyses. B, At least 1 well/plate was also treated with Torin1 to induce autophagy and confirm that a lack of aggregates could be scored on the particular plate. C, Typical data for those infections scored as primary “hits” for clones able to decrease aggregate load in cardiomyocytes subjected to the proteotoxic stimulus of CryABR120G expression. Arrows point to examples of regions that the algorithm scored as positive for aggregates. Shown is a plate of mixed cardiomyocytes and fibroblasts in which the “Control” panel was infected with Ad-CryABR120G but empty lentivirus. Shown in the “shRNA” panels are similar wells: an example of a well scored as a hit is shown. Note the staining across the different sampled areas of the well (top) and a striking decrease in aggregate content (yellow staining). Cardiomyocytes are stained red (TnI antibody) and nuclei blue (DAPI).