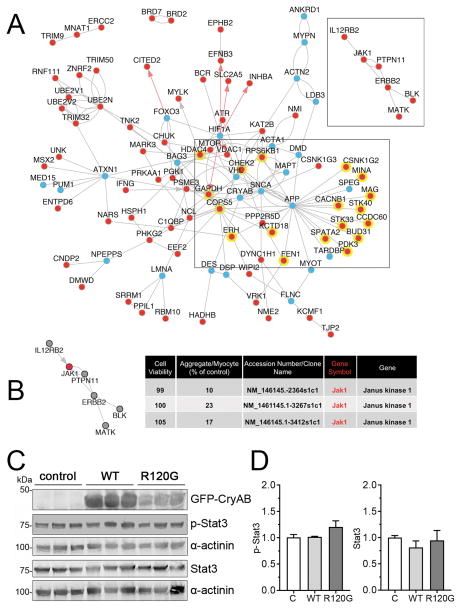
Figure 4. Systems biology prioritization of proteotoxicity regulators and Jak1 signaling.
We first used the OMIM and EntrezGene databases to identify proteotoxicity-annotated genes, and then extended the identified gene sets to related biological processes using ToppGene and AltAnalyze. AltAnalyze NetPerspective network combining proteotoxicity related genes and primary hits with shared direct protein-protein (BIOGRID, grey lines), pathway (WikiPathways, KEGG, grey arrows) or transcriptional regulatory (PAZAR database, red arrows) were visualized. A, A large proportion of primary hits (red dots) have reported interactions with known proteotoxicity genes (blue dots), including 18, which interact with amyloid precursor protein (APP): APP interacting genes are highlighted with a yellow background. A side cluster emerged centered about Jak1 (shown). B, Jak1 knockdown resulted in >75% reduction in aggregates using 3 individual shRNAs directed at the same gene transcript, with no effect to cell viability. A side cluster emerged centered about Jak1 (shown). C, NRVMs were transduced with adenovirus encoding either GFP-CryABWT or GFP-CryABR120G or left untransduced (control; C). Five days after transduction, cells were harvested for protein extraction. A, Representative Western blots stained for p-Stat3 and Stat3. α-actinin was used as a loading control. D, p-Stat3 and Stat3 quantification. Data are presented as mean ± SEM with n=5.