Abstract
Background
Fetal alcohol spectrum disorders (FASD) describe many of the well-known neurodevelopmental deficits afflicting children exposed to alcohol in utero. The effects of alcohol on the maternal-fetal interface, especially the placenta, have been less explored. We herein hypothesized that chronic binge alcohol exposure during pregnancy significantly alters the placental protein profile in a rat FASD model.
Methods
Pregnant rats were orogastrically treated daily with alcohol (4.5 g/kg, gestational day (GD) 5–10; 6.0 g/kg, GD 11–19) or 50% maltose dextrin (isocalorically matched pair-fed controls). On GD 20, placentae were collected, flash frozen, and stored until tissues were homogenized. Protein lysates were denatured, reduced, captured on a 10 kDa spin filter and digested. Peptides were eluted, reconstituted, and analyzed by a Q Exactive™ Hybrid Quadrupole-Orbitrap™ mass spectrometer.
Results
Mass spectrometry analysis identified 2,285 placental proteins based on normalized spectral counts and 2000 proteins by intensity based absolute quantification. 45 placental proteins were significantly (P<0.05) altered by gestational alcohol exposure by both quantification approaches. These included proteins directly related to alcohol metabolism; specific isoforms of alcohol dehydrogenase and aldehyde dehydrogenase were upregulated in the alcohol group. Ingenuity analysis identified ethanol degradation as the most significantly altered canonical pathway in placenta, and fetal/organ development as most altered function, with increased risk for metabolic, neurological, and cardiovascular diseases. Physiologic roles of the significantly altered proteins were related to early pregnancy adaptations, implantation, gestational diseases, fetal organ development, neurodevelopment, and immune functions.
Conclusions
We conclude that the placenta is a valuable organ not only to understand FASD etiology but it may also serve as a diagnostic tool to identify novel biomarkers for detecting the outcome of fetal alcohol exposure. Placental mass spectrometry analysis can offer sophisticated insights into identifying alcohol metabolism-related enzymes and regulators of fetal development.
Keywords: Alcohol, FASD, Teratology
Introduction
Gestational alcohol exposure may result in irreversible fetal damage manifested as a range of physical, physiological, behavioral, and intellectual deficits which are classified under the umbrella term Fetal Alcohol Spectrum Disorders (FASD; Sokol et al., 2003). Prenatal alcohol exposure affects nearly all fetal organ systems including cardiac, vascular, endocrine, neurobehavioral, and uteroplacental systems (Burd et al., 2007a, Ren et al., 2002, Ramadoss and Magness, 2012, Parnell et al., 2007, Warren et al., 2011, Riley and McGee, 2005, Lewis et al., 2015, Gautam et al., 2015, Sawant et al., 2014, Gareri et al., 2009, Gundogan et al., 2008). Fetal Alcohol Syndrome (FAS), the most severe form of FASD, is identifiable in infancy by craniofacial abnormalities and neurobehavioral deficits (Murawski et al., 2015, Jones and Smith, 1973). In contrast, less severe forms of FASD are typically not diagnosed until a child exhibits learning and behavioral problems in elementary school, thereby delaying therapeutic intervention (May et al., 2014). In the United States, 1 in 10 pregnant women consume alcohol and 1 in 33 pregnant women report binge drinking in the past 30 days (Tan et al., 2015), and it is estimated that up to 1 in 20 school children have FASD (May et al., 2009). FASD diagnosis relies on self-reported alcohol consumption during pregnancy, which is often an unreliable and inaccurate method due to social stigma and the fear of litigation (Murawski et al., 2015, Del Boca and Darkes, 2003). Therefore, specific, sensitive and objective methods for detection of in utero alcohol exposure are urgently needed.
Alcohol consumption can be quantified via several diagnostic methods; however, currently no clinical test exists for gestational alcohol exposure or FASD. Studies conducted in men, nonpregnant/pregnant women, and animal models have described numerous markers for alcohol consumption which range in sensitivity, specificity, and window of detection. Candidate biomarkers for alcohol consumption include alcohol metabolites such as acetaldehyde adducts like hemoglobin-associated acetaldehyde (HAA), phosphatidylethanol (PEth), ethyl glucuronide (EtG), ethyl sulfate (EtS), and fatty acid ethyl esters (FAEEs), as well as markers of alcohol-mediated pathology like gamma-glutamyl transferase (GGT), aspartate aminotransferase (AST), alanine aminotransferase (ALT), and mean corpuscular volume (MCV; Joya et al., 2012, Cook, 2003, Peterson, 2004). Specifically in the FASD field, biomarkers are identified utilizing biological samples from the mother like serum/whole blood or directly from the offspring including neonatal hair, meconium, and dried umbilical cord blood spots (Sanvisens et al., 2016, Bakhireva et al., 2013). The placenta is a fetal tissue which is present from implantation to parturition and is highly under-investigated despite its potential as a non-invasive diagnostic medium available at delivery.
The placenta allows for exchange of oxygen, nutrients, waste products, hormones, and antibodies between mother and the developing fetus throughout pregnancy (Sood et al., 2006). The placenta forms at the time of implantation (Bowman and Kennedy, 2014), is delivered after birth, and serves as a medium to obtain valuable information about the health of a pregnancy (Sood et al., 2006, Shukla et al., 2011). Pathological changes in the placenta may be induced by alcohol and are heavily associated with fetal morbidity (Altshuler, 1984), preeclampsia (Walsh, 1985, Pineles et al., 2007), and increased risk for chronic adult diseases such as type 2 diabetes and cardiovascular disease (Jansson and Powell, 2007, Belkacemi, 2011). A growing body of work suggests gestational alcohol-induced maternal-fetal interface adaptations play a critical role in FASD pathogenesis, yet this area of focus remains largely unexplored. Currently, only a limited understanding of the negative effects of gestational alcohol consumption on the placenta exists. In animal models, alcohol alters placental gene expression, indicating the potential of this organ as an effective tool in detecting prenatal alcohol exposure (Shukla et al., 2011, Rosenberg et al., 2010, Balaraman et al., 2014). Thus, we hypothesize that gestational alcohol exposure will significantly alter the placental protein profile in a FASD rat model. Further, we believe that the alcohol-induced placental protein profile will provide pivotal insights into the underlying FASD mechanisms, offer insights into identification of non-invasive FASD biomarkers, and identify targets for developing novel therapeutic intervention.
Materials and Methods
Animals
All experimental procedures were in accordance with National Institutes of Health guidelines (NIH Publication No. 85–23, revised 1996) with approval by the Animal Care and Use Committee at Texas A&M University. Pregnant Sprague Dawley rats were housed in a temperature-controlled room (23°C) with a 12:12-hour light–dark cycle. Rats were assigned to a pair-fed control group (n= 6) or an alcohol treatment group (n = 6). The alcohol group received a dosing regimen of 22.5% (wt/v) ethanol (4.5 g/kg) between GD 5–10. The pair-fed controls were isocalorically matched to alcohol rats by dosing with maltose dextrin to account for calories from alcohol, and the amount of diet consumed by the pair-fed animals was matched with the alcohol-fed animals. Animals received a once daily orogastric gavage in a binge paradigm. Animals were sacrificed on GD 20, one day after the last alcohol exposure. There was no significant difference in the maternal weight between the pair-fed control and alcohol-fed dams on GD 20 (Pair-fed control, 309 ± 8 g; Alcohol, 308 ± 15 g). Fetal weight was significantly decreased in the alcohol group (2.12 ± 0.11 g), compared with that in the pair-fed control (2.53 ± 0.06 g). Placental tissue was isolated, serially washed in phosphate buffered saline (PBS) to remove any residual blood, centrifuged, flash frozen, and stored at −80°C.
Sample Preparation
As previously described (Berger et al., 2015), samples were thawed on ice, washed five times in ice cold PBS, trimmed and shredded using a scalpel, and immediately transferred into ice cold PBS. After vortexing samples for 10 sec, tissues were collected by centrifugation at 4,000 rpm for 10 sec. PBS was removed and this procedure was repeated five times. Blood-free tissue was then collected with 500 μl radioimmunoprecipitation assay (RIPA) buffer (1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, 158 mM NaCl, 10 mM Tris-HCl, pH 7.6, 1 mM PMSF, and protease inhibitor cocktail) and directly transferred to a glass homogenizer for homogenization. For improved cell lysis, the homogenized samples were additionally incubated on ice for 30 min.
Protein Concentration Determination
Protein concentration of the lysates were determined by using the Bradford Assay (Bradford, 1976; Bio-Rad DC™ Protein Assay) following the manufacturer’s protocol. The standard curve was established using a stock solution of 20 mg/ml bovine serum albumin (BSA) and final concentrations of 0.25 mg/ml, 0.5 mg/ml, 1 mg/ml, 1.5 mg/ml and 2.0 mg/ml. After incubation at room temperature, analysis was performed via microplate spectrophotometer (Bio-Rad Model 680) at a wavelength of 595 nm, as previously described (Berger et al., 2015).
Filter assisted sample preparation (FASP)
The filter assisted sample preparation method was carried out as previously described (Wisniewski et al., 2009, Berger et al., 2015). In short: Proteins were first denatured and reduced by adding 100 μl sample to 100 μg urea (8M) supplemented with 20 μl dithiothreitol (DTT, 100 mM in 1 M Tris/HCl pH 8.5). A nominal protein content of 100 μg was used for analysis. After alkylation of reduced cysteine side chains with 50 mM iodoacetic acid (IAA; final concentration), denatured proteins were captured on a 10 kDa molecular weight cut-off (MWCO) spin filter (MRCPRT010, Millipore) and washed twice with 50 mM ammonium bicarbonate (ABC). Protein digestion was performed with sequencing grade trypsin (V5111, Promega) at a nominal enzyme to substrate ratio of 1:50. After incubation overnight with 100 μl digestion buffer (trypsin in 50 mM ABC), resulting peptides were eluted with 300 μl 0.5 M sodium chloride (NaCl). Peptide elutes were desalted with reversed, phase-based, TARGA C-18 spin tips (SEMSS18R, Nest Group) prior to LC-MS/MS analysis. Lyophilized samples were stored at −20°C.
LC-MS/MS Analysis
A combination of liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS), as previously described (Berger et al., 2015), was implemented for the analysis of placental tissue. In brief, following the ultra-performance liquid chromatography (UPLC), the first stage of MS (MS1) produced precursor ion fragmentation, and in the second stage MS (MS2) separated and detected the produced fragmented ions. Peptides were reconstituted in loading buffer (5% acetonitrile [ACN; v/v], 5% formic acid [FA; v/v]). The prepared samples were analyzed using a nanoflow UPLC system (EK425, Sciex) with an in-house packed column (75 μm × 15 cm; AQUA C18/3 μm from Michrom Bioresource) coupled online to a Q Exactive ™ Hybrid Quadrupole-Orbitrap™ mass spectrometer (MS; Thermo Fisher Scientific), as previously described (Berger et al., 2015). Tryptic digests (~1.5 μg) were separated by a linear gradient from 93% buffer A (0.2% FA in HPLC water)/7% buffer B (0.2% FA in ACN) to 70% buffer A/30% buffer B within 120 min at a flowrate of 400 nl/min. The MS was operated in data-dependent TOP10 mode with the following settings: resolution for MS1 scan: 70,000 at 200 Th; lock mass: 445.120025 Th; resolution for MS2 scan: 17,500 at 200 Th; isolation width: 1.6 m/z; NCE 27; underfilll ratio: 1%; charge state exclusion: unassigned, 1, > 6; dynamic exclusion: 20 sec.
Data Analysis
Acquired MS raw files (RAW) were analyzed using MaxQuant (Cox and Mann, 2008, Berger et al., 2015; version 1.5.2.8). Briefly, the acquired RAW files were loaded into MaxQuant and searched against the rat UniProtKB database. For quantification, ‘intensity based absolute quantification’ (iBAQ) and ‘label-free quantification’ (LFQ) were selected. Fixed modifications were set to Carbamidomethyl (C) and variable modifications to Acetyl (N-Term), Oxidation (M), and Phospho (STY), as digesting enzyme ‘trypsin’ was specified. Otherwise, default settings (by MaxQuant) were used for the analysis: trypsin with up to two missed cleavages; mass tolerance for the first search: 0.07 Da; main search: 0.006 Da. Additionally, RAW files were converted into Mascot Generic Files (MGF) using an in-house script (ms2preproc, version 2009-01). The generated MGF files were searched against the rat database (containing all sequences in a reversed order as decoy) using Mascot (Matrix Science; version 2.3). The following parameters were used for the search: ‘enzyme’ was set to ‘trypsin’, two missed cleavages were allowed, peptide tolerance was set to 10 ppm and the MS/MS tolerance to 20 mmu, only 2+, 3+ and 4+ charged ions were allowed, search was performed in Target-Decoy mode. Mascot search result files (DAT files) were loaded to Scaffold (Proteome Software Inc., version 4.3.2) for further analysis. General contaminants were not included in the subsequent data analysis. Protein abundances were determined using a normalized spectral count method as well as protein iBAQ. Identified proteins were evaluated by a blinded observer using Ingenuity Pathway Analysis to interpret the proteomic data.
Statistics
Difference in the mean protein abundance between pair-fed control and alcohol treatment groups was analyzed using a Student’s t-test. Data are presented as Mean + SEM. Significance was established a priori at P < 0.05.
Results
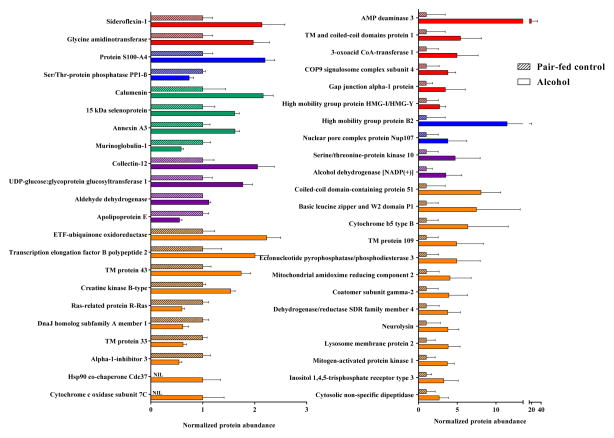
Two approaches were used to quantify the placental proteins identified by mass spectrometry: a total of 2,285 proteins with greater than 95% probability and a minimum of 2 peptides per protein were identified and quantified by normalized spectral counts method with Mascot and Scaffold, and 2000 proteins by iBAQ analysis with MaxQuant. A list of identified proteins is provided in the Supplemental Information. 321 of the proteins detected by the normalized spectral method and 262 of the proteins identified by the iBAQ method were significantly (P < 0.05) altered by gestational alcohol exposure. A stringent approach was utilized to validate the proteins significantly altered in response to alcohol: proteins identified by both iBAQ and normalized spectral counts methods, met validation criteria: 45 placental proteins were identified by both quantification processes as significantly different in pair-fed controls and alcohols (Figure 1).
Figure 1.
Mass spectrometry analysis of alcohol-induced alteration of placental proteins. 45 proteins were identified as significantly different (P < 0.05) between the pair-fed control and alcohol groups when quantified by both normalized spectral counts method with Mascot/Scaffold, and by iBAQ analysis with MaxQuant. Colors depict protein function categorization: Red – Gestational diseases, fetal development; Blue – Neurodevelopment; Green – Implantation; Purple – Alcohol metabolism, nutrition, and immune function; and Orange – Cellular function. Data are represented as mean ± SEM.
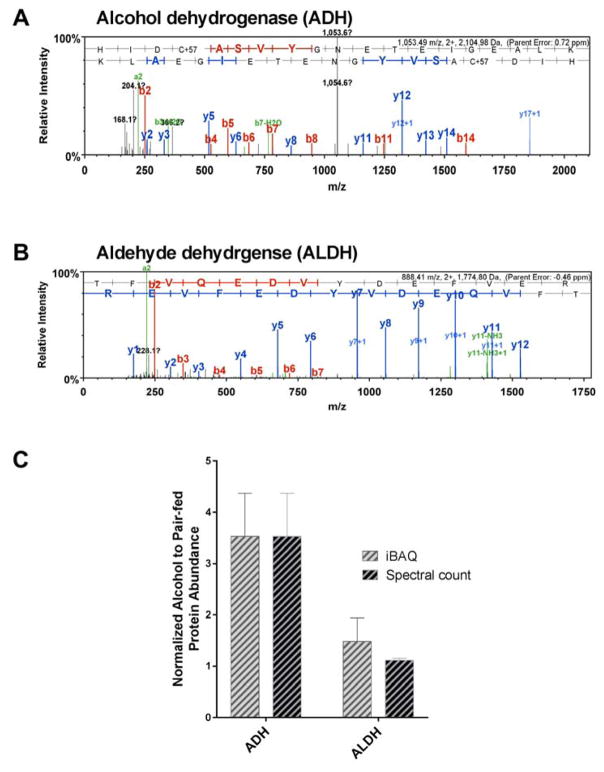
Interestingly, proteins directly related to alcohol metabolism (Crabb et al., 2004) were upregulated in the alcohol group, including both alcohol dehydrogenase NADP(+) (ADH) and aldehyde dehydrogenase, mitochondrial (ALDH). The representative MS/MS spectra illustrated in Figure 2 depict a nearly complete y and b ion series assignment for ADH (parent mass error: 0.72 ppm; Figure 2A) and ALDH (parent mass error: −0.46 ppm; Figure 2B). The b ion series identified peptide fragments which extend from the amino (N)-terminus, whereas, the y ion series identified the peptide fragments extending from the carboxyl (C)-terminus. Peak height identified the MS signal intensity, and the distance between the peaks was used to construct the sequences of the identified peptide. The mean alcohol to pair-fed protein abundance of ADH was determined to be 3.53 by iBAQ, and 3.53 by normalized spectral count, and the mean alcohol to pair-fed protein abundance for ALDH was 1.49 by iBAQ, and 1.12 by normalized spectral count (Figure 2C).
Figure 2.
Proteins directly related to alcohol metabolism were upregulated in the alcohol group, which included both alcohol dehydrogenase NADP(+) (ADH) and aldehyde dehydrogenase, mitochondrial (ALDH). One representative MS/MS spectra of (A) ADH (parent mass error: 0.72 ppm) and (B) ALDH (parent mass error: −0.46 ppm) from alcohol-exposed placenta illustrate the ions detected following protein fragmentation. Each vertical peak identifies the relative abundance of the ion and the corresponding peptide sequence, based on the mass to charge ratio (m/z). The distance between the peaks is used to construct the sequences of the identified peptide following protein fragmentation. The b ion series (red) identifies peptide fragments extending from the amino (N)-terminus, and the y ion series (blue) identifies peptide fragments extending from the carboxyl (C)-terminus. (C) The mean alcohol to pair-fed protein abundance of ADH was determined to be 3.53 by iBAQ, and 3.53 by normalized spectral count, and the mean alcohol to pair-fed protein abundance for ALDH was 1.49 by iBAQ, and 1.12 by normalized spectral count.
Ingenuity Pathway Analysis identified canonical pathways, diseases, and physiological systems development and functions significantly altered in the placenta following gestational alcohol exposure (Table 1). Ethanol degradation, noradrenaline and adrenaline degradation, and synaptic long term potentiation were the top canonical pathways altered following gestational alcohol exposure. Analysis also revealed increased risk for metabolic, neurological, and cardiovascular diseases following gestational alcohol exposure. Importantly, the most altered physiologic systems were directly related to fetal organ and embryo development, hair and skin development, and auditory, vestibular, renal, and urological systems function.
Table 1.
Ingenuity analysis identified ethanol degradation as the most significantly altered canonical pathway in the placenta with significantly increased risk for metabolic, neurological, and cardiovascular diseases. The most altered physiologic systems were directly related to fetal organ and embryo development.
| INGENUITY Pathway Analysis | |
|---|---|
| Top Canonical Pathways | P Value |
| Ethanol Degradation II | 4.01×10−5 |
| Noradrenaline and Adrenaline Degradation | 5.09×10−5 |
| Synaptic Long Term Potentiation | 6.33×10−5 |
| Top Diseases | |
| Metabolic Disease | 1.43×10−2 – 2.14×10−5 |
| Cardiovascular Disease | 1.76×10−2 – 2.84×10−5 |
| Neurological Disease | 1.79×10−2 – 1.42×10−5 |
| Physiological System Development and Function | |
| Renal and Urological System Development and Function | 1.25×10−2 – 1.41×10−4 |
| Auditory and Vestibular System Development and Function | 1.79×10−2 – 3.27×10−4 |
| Embryonic Development | 1.79×10−2 – 1.13×10−5 |
| Organismal Development | 1.79×10−2 – 4.72×10−5 |
| Hair and Skin Development and Function | 3.60×10−3 – 1.13×10−5 |
We classified the significantly altered proteins based on their physiological functions (Figure 3). A number of proteins related to early pregnancy adaptations and implantation were downregulated, such as 15 kDa selenoprotein and calumenin. Proteins involved in preeclampsia, gestational diseases, and fetal organ development, including AMP deaminase 3 and glycine amidinotransferase, were also downregulated. Proteins involved in neurodevelopment were also altered by alcohol: high mobility group protein B was downregulated, and serine/threonine-protein phosphatase PP1-beta was upregulated. Importantly, proteins essential for alcohol metabolism and nutrition, such as alcohol dehydrogenase and aldehyde dehydrogenase, were upregulated in the alcohol group. Proteins involved in immune functions, such as UDP-glucose:glycoprotein glycosyltransferase and collectin-12, were downregulated in the placenta following gestational alcohol exposure.
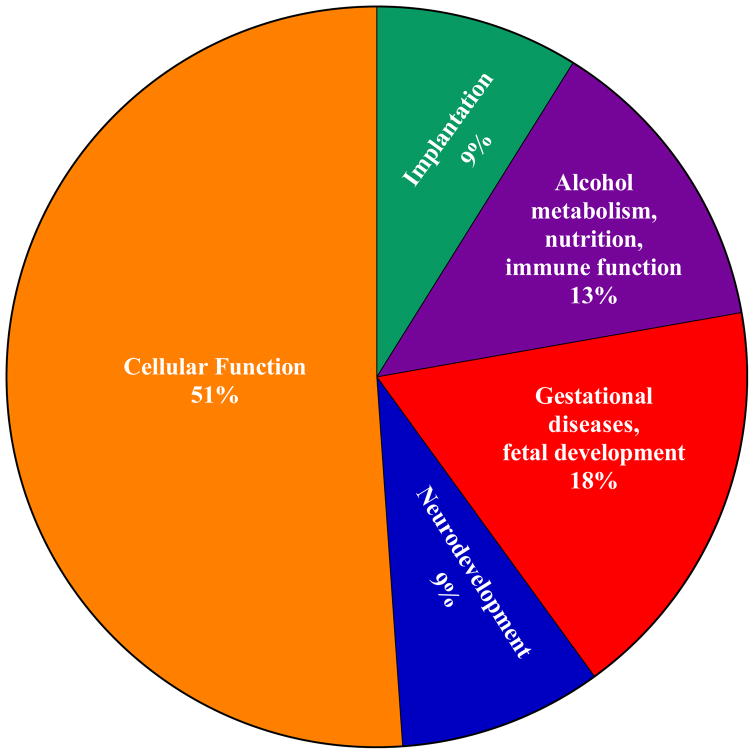
Figure 3.
Categorization of proteins significantly altered by gestational alcohol exposure based on their physiologic function during pregnancy. Specifically, 51% of the proteins were associated with general cellular functions, 18% with gestational diseases and fetal organ development, 13% with alcohol metabolism, nutrition and immune function, 9% with implantation, binding, and gestational adaptations, and 9% with fetal neurodevelopment.
Discussion
To the best of our knowledge, this is the first study to investigate the placental protein profile in the FASD field. We herein demonstrate that chronic binge alcohol exposure has specific and substantial effects on the placenta; of the more than 2000 placental proteins identified, 45 proteins were significantly altered in the alcohol treatment group compared to the pair-fed control group when analyzed by both normalized spectral count and the iBAQ approaches. Ethanol degradation was the most significantly altered canonical pathway in the placenta. Placental proteins altered by alcohol indicated increased risk for metabolic, neurological, and cardiovascular diseases. Our data also demonstrate that the placental proteins may act as a window into fetal/organ development in the FASD rat model.
Alcohol freely disperses from maternal circulation across the placenta into the fetal compartment, producing fetal BACs and amniotic fluid concentrations equivalent to maternal BACs, yet alcohol remains for a longer duration within the amniotic fluid (Nava-Ocampo et al., 2004). The placenta has multiple functions during pregnancy (Burd et al., 2007b) and previous studies have shown that alcohol (1) induces placental vasoconstriction, which persists for the duration of exposure (Taylor et al., 1994) and adversely impacts nutrient and oxygen delivery to the fetus (Burd et al., 2007b); (2) alters placental metabolism by decreasing glucose utilization (Rice et al., 1986, Burd et al., 2007b); (3) disrupts endocrine function by inhibiting growth factor production (Karl and Fisher, 1994); and (4) activates an immune response by upregulating cytokine expression (Svinarich et al., 1998). Together these alcohol-induced changes impact multiple maternal placenta-associated syndromes; women who drank during pregnancy were more likely to develop placental abruption, have an infant that is small for gestational age, and have a preterm infant when compared with mothers who abstain (Salihu et al., 2011). The mechanism(s) underlying alcohol-induced placental pathogenesis requires further study.
We analyzed the physiological functions of placental proteins altered by gestational alcohol exposure and found they had critical roles in alcohol metabolism, pregnancy, placental function, and fetal development. Ethanol degradation was the most significantly altered canonical pathway in the placenta. ADH and ALDH have integral roles in alcohol metabolism (Tawa et al., 2016), and were determined to be upregulated by mass spectrometry analysis. Proteins involved in alcohol-related disease were also upregulated such as sideroflexin-1 (Rosenberg et al., 2010), which is linked to alcoholic hepatotoxicity and iron transport (Kim et al., 2015). The fetal liver is very limited in its ability to metabolize ethanol and additional studies are necessary to further decipher the expression, distribution, and activity of ALDH and ADH in the placenta (Sanchis and Guerri, 1986, Cederbaum, 2012, Pikkarainen and Raiha, 1967, Heller and Burd, 2014). This study demonstrates that ALDH and ADH are expressed in the placenta following chronic alcohol exposure and these enzymes may have the ability to metabolize alcohol. Future studies are warranted to assess the placental metabolic clearance of alcohol. Mass spectrometry analysis determined GATM to be significantly upregulated following gestational alcohol exposure. GATM is involved in the biosynthesis of guanidinoacetate, a precursor of creatine (Item et al., 2001). Diseases associated with GATM mutations have resulted in deficiencies in creatine synthesis which may result in mental and behavioral disorders (Fons and Campistol, 2016). Consequently, GATM may have a critical role in neurodevelopment and its disruption may affect the neuropathogenesis of FASD.
Our data also demonstrate that the placental proteins may act as a window into fetal/organ development in the FASD rat model and they indicate risk for alcohol-induced metabolic, neurological, and cardiovascular diseases. Gestational alcohol exposure altered proteins critical for early pregnancy adaptations and implantation: murinoglobulin-1, secreted from uterine stromal cells and involved in trophoblast invasion (Esadeg et al., 2003) was downregulated, while 15 kDa selenoprotein (Mistry et al., 2012) and calumenin (Mahnke-Zizelman et al., 1997) were upregulated. Proteins related to preeclampsia, gestational diseases, and fetal organ development were also upregulated, including AMP deaminase 3, a regulator of skeletal muscle and liver development (Mahnke-Zizelman et al., 1997), and glycine amidinotransferase, critical for embryonic and central nervous system development (McMinn et al., 2006). Furthermore, proteins involved in neurodevelopment were dysregulated following gestational alcohol exposure: high mobility group protein B (Abraham et al., 2013) was upregulated, while serine/threonine-protein phosphatase PP1-beta (Chiappetta et al., 1996) was downregulated. FASD-induced infection susceptibility may be imprinted during early development, as we identified proteins involved in the immune response were upregulated, such as UDP-glucose:glycoprotein glycosyltransferase (Zhang et al., 2011), collectin-12 (Ohtani et al., 2012), and serine/threonine-protein kinase 10 (Yamamoto et al., 2011). In summary, our data demonstrate that alcohol modifies protein profiles comprising early pregnancy and implantation, preeclampsia, gestational diseases, fetal organ development, and immune functions.
In conclusion, the placenta is unique in that it subsists exclusively from implantation until birth, and is subjected to nearly identical environmental exposures as the developing fetus. Additional studies are warranted for further validation and for assessment of placental metabolic clearance of alcohol. We anticipate that placental alterations following gestational alcohol exposure may accurately reflect pathological changes occurring within the fetus. Thus, the placenta may serve as a medium for early detection and subsequent intervention for deficits where detection may be invasive, inaccessible, or not apparent until later in life, as is the case with many alcohol-induced neurobehavioral abnormalities. The findings of this study build on earlier reports in the field and help delineate how placental protein dysregulation may provide insights into FASD phenotypes.
Supplementary Material
Acknowledgments
Grants: NIH AA19446, AA23520, AA23035, Texas A&M Tier One Program (JR)
We appreciate the valuable technical assistance from Marcus Orzabal, Shawn Erin, Brian Hickner, and Thomas Tigner.
Footnotes
Disclosures: None
References
- ABRAHAM AB, BRONSTEIN R, CHEN EI, KOLLER A, RONFANI L, MALETIC-SAVATIC M, TSIRKA SE. Members of the high mobility group B protein family are dynamically expressed in embryonic neural stem cells. Proteome Sci. 2013;11:18. doi: 10.1186/1477-5956-11-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ALTSHULER G. Chorangiosis. An important placental sign of neonatal morbidity and mortality. Arch Pathol Lab Med. 1984;108:71–4. [PubMed] [Google Scholar]
- BAKHIREVA LN, SAVICH RD, RAISCH DW, CANO S, ANNETT RD, LEEMAN L, GARG M, GOFF C, SAVAGE DD. The feasibility and cost of neonatal screening for prenatal alcohol exposure by measuring phosphatidylethanol in dried blood spots. Alcohol Clin Exp Res. 2013;37:1008–15. doi: 10.1111/acer.12045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BALARAMAN S, LUNDE ER, SAWANT O, CUDD TA, WASHBURN SE, MIRANDA RC. Maternal and neonatal plasma microRNA biomarkers for fetal alcohol exposure in an ovine model. Alcohol Clin Exp Res. 2014;38:1390–400. doi: 10.1111/acer.12378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BELKACEMI L, NELSON MD, DESAI M, ROSS MG. Maternal Undernutrition and Fetal Programming: Role of the Placenta. In: KAY HH, NELSON MD, WANG Y, editors. The Placenta: From Development to Disease. Blackwell Publishing Ltd; 2011. [Google Scholar]
- BERGER ST, AHMED S, MUNTEL J, CUEVAS POLO N, BACHUR R, KENTSIS A, STEEN J, STEEN H. MStern Blotting-High Throughput Polyvinylidene Fluoride (PVDF) Membrane-Based Proteomic Sample Preparation for 96-Well Plates. Mol Cell Proteomics. 2015;14:2814–23. doi: 10.1074/mcp.O115.049650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BOWMAN ZS, KENNEDY AM. Sonographic appearance of the placenta. Curr Probl Diagn Radiol. 2014;43:356–73. doi: 10.1067/j.cpradiol.2014.05.006. [DOI] [PubMed] [Google Scholar]
- BRADFORD MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–54. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- BURD L, DEAL E, RIOS R, ADICKES E, WYNNE J, KLUG MG. Congenital heart defects and fetal alcohol spectrum disorders. Congenit Heart Dis. 2007a;2:250–5. doi: 10.1111/j.1747-0803.2007.00105.x. [DOI] [PubMed] [Google Scholar]
- BURD L, ROBERTS D, OLSON M, ODENDAAL H. Ethanol and the placenta: A review. J Matern Fetal Neonatal Med. 2007b;20:361–75. doi: 10.1080/14767050701298365. [DOI] [PubMed] [Google Scholar]
- CEDERBAUM AI. Alcohol metabolism. Clin Liver Dis. 2012;16:667–85. doi: 10.1016/j.cld.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHIAPPETTA G, AVANTAGGIATO V, VISCONTI R, FEDELE M, BATTISTA S, TRAPASSO F, MERCIAI BM, FIDANZA V, GIANCOTTI V, SANTORO M, SIMEONE A, FUSCO A. High level expression of the HMGI (Y) gene during embryonic development. Oncogene. 1996;13:2439–46. [PubMed] [Google Scholar]
- COOK JD. Biochemical markers of alcohol use in pregnant women. Clin Biochem. 2003;36:9–19. doi: 10.1016/s0009-9120(02)00424-1. [DOI] [PubMed] [Google Scholar]
- COX J, MANN M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–72. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- CRABB DW, MATSUMOTO M, CHANG D, YOU M. Overview of the role of alcohol dehydrogenase and aldehyde dehydrogenase and their variants in the genesis of alcohol-related pathology. Proc Nutr Soc. 2004;63:49–63. doi: 10.1079/pns2003327. [DOI] [PubMed] [Google Scholar]
- DAYA D, SABET L. The use of cytokeratin as a sensitive and reliable marker for trophoblastic tissue. Am J Clin Pathol. 1991;95:137–41. doi: 10.1093/ajcp/95.2.137. [DOI] [PubMed] [Google Scholar]
- DEL BOCA FK, DARKES J. The validity of self-reports of alcohol consumption: state of the science and challenges for research. Addiction. 2003;98(Suppl 2):1–12. doi: 10.1046/j.1359-6357.2003.00586.x. [DOI] [PubMed] [Google Scholar]
- ESADEG S, HE H, PIJNENBORG R, VAN LEUVEN F, CROY BA. Alpha-2 macroglobulin controls trophoblast positioning in mouse implantation sites. Placenta. 2003;24:912–21. doi: 10.1016/s0143-4004(03)00148-6. [DOI] [PubMed] [Google Scholar]
- FONS C, CAMPISTOL J. Creatine Defects and Central Nervous System. Semin Pediatr Neurol. 2016;23:285–289. doi: 10.1016/j.spen.2016.11.003. [DOI] [PubMed] [Google Scholar]
- GARERI J, BRIEN J, REYNOLDS J, KOREN G. Potential role of the placenta in fetal alcohol spectrum disorder. Paediatr Drugs. 2009;11:26–9. doi: 10.2165/0148581-200911010-00010. [DOI] [PubMed] [Google Scholar]
- GAUTAM P, LEBEL C, NARR KL, MATTSON SN, MAY PA, ADNAMS CM, RILEY EP, JONES KL, KAN EC, SOWELL ER. Volume changes and brain-behavior relationships in white matter and subcortical gray matter in children with prenatal alcohol exposure. Hum Brain Mapp. 2015;36:2318–29. doi: 10.1002/hbm.22772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GUNDOGAN F, ELWOOD G, LONGATO L, TONG M, FEIJOO A, CARLSON RI, WANDS JR, DE LA MONTE SM. Impaired placentation in fetal alcohol syndrome. Placenta. 2008;29:148–57. doi: 10.1016/j.placenta.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HELLER M, BURD L. Review of ethanol dispersion, distribution, and elimination from the fetal compartment. Birth Defects Res A Clin Mol Teratol. 2014;100:277–83. doi: 10.1002/bdra.23232. [DOI] [PubMed] [Google Scholar]
- ITEM CB, STOCKLER-IPSIROGLU S, STROMBERGER C, MUHL A, ALESSANDRI MG, BIANCHI MC, TOSETTI M, FORNAI F, CIONI G. Arginine:glycine amidinotransferase deficiency: the third inborn error of creatine metabolism in humans. Am J Hum Genet. 2001;69:1127–33. doi: 10.1086/323765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JANSSON T, POWELL TL. Role of the placenta in fetal programming: underlying mechanisms and potential interventional approaches. Clin Sci (Lond) 2007;113:1–13. doi: 10.1042/CS20060339. [DOI] [PubMed] [Google Scholar]
- JOHNSON GA, BAZER FW, JAEGER LA, KA H, GARLOW JE, PFARRER C, SPENCER TE, BURGHARDT RC. Muc-1, integrin, and osteopontin expression during the implantation cascade in sheep. Biol Reprod. 2001;65:820–8. doi: 10.1095/biolreprod65.3.820. [DOI] [PubMed] [Google Scholar]
- JONES KL, SMITH DW. Recognition of the fetal alcohol syndrome in early infancy. Lancet. 1973;302:999–1001. doi: 10.1016/s0140-6736(73)91092-1. [DOI] [PubMed] [Google Scholar]
- JOYA X, FRIGULS B, ORTIGOSA S, PAPASEIT E, MARTINEZ SE, MANICH A, GARCIA-ALGAR O, PACIFICI R, VALL O, PICHINI S. Determination of maternal-fetal biomarkers of prenatal exposure to ethanol: a review. J Pharm Biomed Anal. 2012;69:209–22. doi: 10.1016/j.jpba.2012.01.006. [DOI] [PubMed] [Google Scholar]
- KARL P, FISHER S. Chronic Ethanol Exposure Inhibits Insulin and IGF-1 Stimulated Amino Acid Uptake in Cultured Human Placental Trophoblasts. Alcoholism: clinical and experimental research. 1994;18:942–946. doi: 10.1111/j.1530-0277.1994.tb00063.x. [DOI] [PubMed] [Google Scholar]
- KIM DH, LEE EM, DO SH, JEONG DH, JEONG KS. Changes of the Cytoplasmic Proteome in Response to Alcoholic Hepatotoxicity in Rats. Int J Mol Sci. 2015;16:18664–82. doi: 10.3390/ijms160818664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEWIS CE, THOMAS KG, DODGE NC, MOLTENO CD, MEINTJES EM, JACOBSON JL, JACOBSON SW. Verbal learning and memory impairment in children with fetal alcohol spectrum disorders. Alcohol Clin Exp Res. 2015;39:724–32. doi: 10.1111/acer.12671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MAHNKE-ZIZELMAN DK, D’CUNHA J, WOJNAR JM, BROGLEY MA, SABINA RL. Regulation of rat AMP deaminase 3 (isoform C) by development and skeletal muscle fibre type. Biochem J. 1997;326(Pt 2):521–9. doi: 10.1042/bj3260521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MAY PA, BAETE A, RUSSO J, ELLIOTT AJ, BLANKENSHIP J, KALBERG WO, BUCKLEY D, BROOKS M, HASKEN J, ABDUL-RAHMAN O, ADAM MP, ROBINSON LK, MANNING M, HOYME HE. Prevalence and characteristics of fetal alcohol spectrum disorders. Pediatrics. 2014;134:855–66. doi: 10.1542/peds.2013-3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MAY PA, GOSSAGE JP, KALBERG WO, ROBINSON LK, BUCKLEY D, MANNING M, HOYME HE. Prevalence and epidemiologic characteristics of FASD from various research methods with an emphasis on recent in-school studies. Dev Disabil Res Rev. 2009;15:176–92. doi: 10.1002/ddrr.68. [DOI] [PubMed] [Google Scholar]
- MCMINN J, WEI M, SCHUPF N, CUSMAI J, JOHNSON EB, SMITH AC, WEKSBERG R, THAKER HM, TYCKO B. Unbalanced placental expression of imprinted genes in human intrauterine growth restriction. Placenta. 2006;27:540–9. doi: 10.1016/j.placenta.2005.07.004. [DOI] [PubMed] [Google Scholar]
- MISTRY HD, BROUGHTON PIPKIN F, REDMAN CW, POSTON L. Selenium in reproductive health. Am J Obstet Gynecol. 2012;206:21–30. doi: 10.1016/j.ajog.2011.07.034. [DOI] [PubMed] [Google Scholar]
- MURAWSKI NJ, MOORE EM, THOMAS JD, RILEY EP. Advances in Diagnosis and Treatment of Fetal Alcohol Spectrum Disorders: From Animal Models to Human Studies. Alcohol Res. 2015;37:97–108. [PMC free article] [PubMed] [Google Scholar]
- NAVA-OCAMPO AA, VELAZQUEZ-ARMENTA Y, BRIEN JF, KOREN G. Elimination kinetics of ethanol in pregnant women. Reprod Toxicol. 2004;18:613–7. doi: 10.1016/j.reprotox.2004.02.012. [DOI] [PubMed] [Google Scholar]
- OHTANI K, SUZUKI Y, WAKAMIYA N. Biological functions of the novel collectins CL-L1, CL-K1, and CL-P1. J Biomed Biotechnol. 2012;2012:493945. doi: 10.1155/2012/493945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PARNELL SE, RAMADOSS J, DELP MD, RAMSEY MW, CHEN WJ, WEST JR, CUDD TA. Chronic ethanol increases fetal cerebral blood flow specific to the ethanol-sensitive cerebellum under normoxaemic, hypercapnic and acidaemic conditions: ovine model. Exp Physiol. 2007;92:933–43. doi: 10.1113/expphysiol.2007.038091. [DOI] [PubMed] [Google Scholar]
- PETERSON K. Biomarkers for alcohol use and abuse--a summary. Alcohol Res Health. 2004;28:30–7. [PMC free article] [PubMed] [Google Scholar]
- PIKKARAINEN PH, RAIHA NC. Development of alcohol dehydrogenase activity in the human liver. Pediatr Res. 1967;1:165–8. doi: 10.1203/00006450-196705000-00001. [DOI] [PubMed] [Google Scholar]
- PINELES BL, ROMERO R, MONTENEGRO D, TARCA AL, HAN YM, KIM YM, DRAGHICI S, ESPINOZA J, KUSANOVIC JP, MITTAL P, HASSAN SS, KIM CJ. Distinct subsets of microRNAs are expressed differentially in the human placentas of patients with preeclampsia. Am J Obstet Gynecol. 2007;196:261, e1–6. doi: 10.1016/j.ajog.2007.01.008. [DOI] [PubMed] [Google Scholar]
- RAMADOSS J, MAGNESS RR. Multiplexed digital quantification of binge-like alcohol-mediated alterations in maternal uterine angiogenic mRNA transcriptome. Physiol Genomics. 2012;44:622–8. doi: 10.1152/physiolgenomics.00009.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- REN J, WOLD LE, NATAVIO M, REN BH, HANNIGAN JH, BROWN RA. Influence of prenatal alcohol exposure on myocardial contractile function in adult rat hearts: role of intracellular calcium and apoptosis. Alcohol Alcohol. 2002;37:30–7. doi: 10.1093/alcalc/37.1.30. [DOI] [PubMed] [Google Scholar]
- RICE PA, NESBITT RE, JR, CUENCA VG, ZHANG W, GORDON GB, KIM TJ. The effect of ethanol on the production of lactate, triglycerides, phospholipids, and free fatty acids in the perfused human placenta. Am J Obstet Gynecol. 1986;155:207–11. doi: 10.1016/0002-9378(86)90112-2. [DOI] [PubMed] [Google Scholar]
- RILEY EP, MCGEE CL. Fetal alcohol spectrum disorders: an overview with emphasis on changes in brain and behavior. Exp Biol Med (Maywood) 2005;230:357–65. doi: 10.1177/15353702-0323006-03. [DOI] [PubMed] [Google Scholar]
- ROSENBERG MJ, WOLFF CR, EL-EMAWY A, STAPLES MC, PERRONE-BIZZOZERO NI, SAVAGE DD. Effects of moderate drinking during pregnancy on placental gene expression. Alcohol. 2010;44:673–90. doi: 10.1016/j.alcohol.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SALIHU HM, KORNOSKY JL, LYNCH O, ALIO AP, AUGUST EM, MARTY PJ. Impact of prenatal alcohol consumption on placenta-associated syndromes. Alcohol. 2011;45:73–9. doi: 10.1016/j.alcohol.2010.05.010. [DOI] [PubMed] [Google Scholar]
- SANCHIS R, GUERRI C. Alcohol-metabolizing enzymes in placenta and fetal liver: effect of chronic ethanol intake. Alcohol Clin Exp Res. 1986;10:39–44. doi: 10.1111/j.1530-0277.1986.tb05611.x. [DOI] [PubMed] [Google Scholar]
- SANVISENS A, ROBERT N, HERNANDEZ JM, ZULUAGA P, FARRE M, COROLEU W, SERRA M, TOR J, MUGA R. Alcohol Consumption during Pregnancy: Analysis of Two Direct Metabolites of Ethanol in Meconium. Int J Mol Sci. 2016;17 doi: 10.3390/ijms17030417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAWANT OB, RAMADOSS J, HANKINS GD, WU G, WASHBURN SE. Effects of L-glutamine supplementation on maternal and fetal hemodynamics in gestating ewes exposed to alcohol. Amino Acids. 2014;46:1981–96. doi: 10.1007/s00726-014-1751-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHUKLA PK, SITTIG LJ, ULLMANN TM, REDEI EE. Candidate placental biomarkers for intrauterine alcohol exposure. Alcohol Clin Exp Res. 2011;35:559–65. doi: 10.1111/j.1530-0277.2010.01373.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SOKOL RJ, DELANEY-BLACK V, NORDSTROM B. Fetal alcohol spectrum disorder. JAMA. 2003;290:2996–9. doi: 10.1001/jama.290.22.2996. [DOI] [PubMed] [Google Scholar]
- SOOD R, ZEHNDER JL, DRUZIN ML, BROWN PO. Gene expression patterns in human placenta. Proc Natl Acad Sci U S A. 2006;103:5478–83. doi: 10.1073/pnas.0508035103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SVINARICH DM, DICERBO JA, ZAHER FM, YELIAN FD, GONIK B. Ethanol-induced expression of cytokines in a first-trimester trophoblast cell line. Am J Obstet Gynecol. 1998;179:470–5. doi: 10.1016/s0002-9378(98)70381-3. [DOI] [PubMed] [Google Scholar]
- TAN CH, DENNY CH, CHEAL NE, SNIEZEK JE, KANNY D. Alcohol use and binge drinking among women of childbearing age - United States, 2011–2013. MMWR Morb Mortal Wkly Rep. 2015;64:1042–6. doi: 10.15585/mmwr.mm6437a3. [DOI] [PubMed] [Google Scholar]
- TAWA EA, HALL SD, LOHOFF FW. Overview of the Genetics of Alcohol Use Disorder. Alcohol Alcohol. 2016;51:507–14. doi: 10.1093/alcalc/agw046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TAYLOR SM, HERON AE, CANNELL GR, FLORIN TH. Pressor effect of ethanol in the isolated perfused human placental lobule. Eur J Pharmacol. 1994;270:371–4. doi: 10.1016/0926-6917(94)90015-9. [DOI] [PubMed] [Google Scholar]
- WALSH SW. Preeclampsia: an imbalance in placental prostacyclin and thromboxane production. Am J Obstet Gynecol. 1985;152:335–40. doi: 10.1016/s0002-9378(85)80223-4. [DOI] [PubMed] [Google Scholar]
- WARREN KR, HEWITT BG, THOMAS JD. Fetal alcohol spectrum disorders: research challenges and opportunities. Alcohol Res Health. 2011;34:4–14. [PMC free article] [PubMed] [Google Scholar]
- WISNIEWSKI JR, ZOUGMAN A, NAGARAJ N, MANN M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6:359–62. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- YAMAMOTO N, HONMA M, SUZUKI H. Off-target serine/threonine kinase 10 inhibition by erlotinib enhances lymphocytic activity leading to severe skin disorders. Mol Pharmacol. 2011;80:466–75. doi: 10.1124/mol.110.070862. [DOI] [PubMed] [Google Scholar]
- ZHANG W, WEARSCH PA, ZHU Y, LEONHARDT RM, CRESSWELL P. A role for UDP-glucose glycoprotein glucosyltransferase in expression and quality control of MHC class I molecules. Proc Natl Acad Sci U S A. 2011;108:4956–61. doi: 10.1073/pnas.1102527108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.