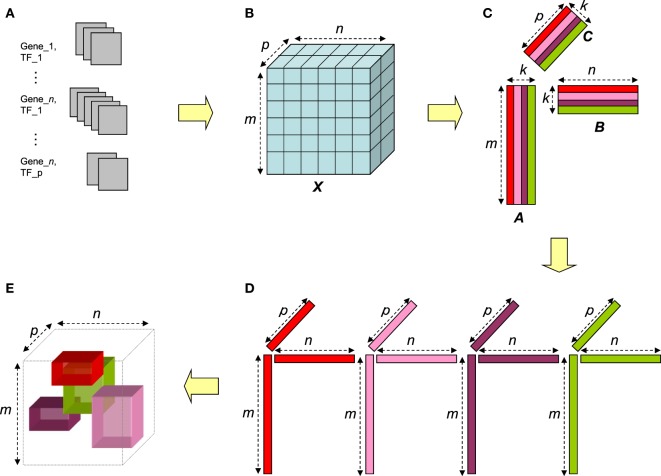
Figure 1.
Overview of the NTF-based procedure. Gene–TF documents (A) are parsed to construct term × gene × TF tensor (B). The tensor is factorized via NTF to generate non-negative factor matrices for terms, genes, and TFs (C). For a given approximation rank k, the entities corresponding to the high magnitude entries in each triad of columns (one from each matrix) (D) can be interpreted as an annotated transcriptional module (ATM) comprising of genes and TFs functionally annotated by the corresponding terms (E).