Fig. 1.
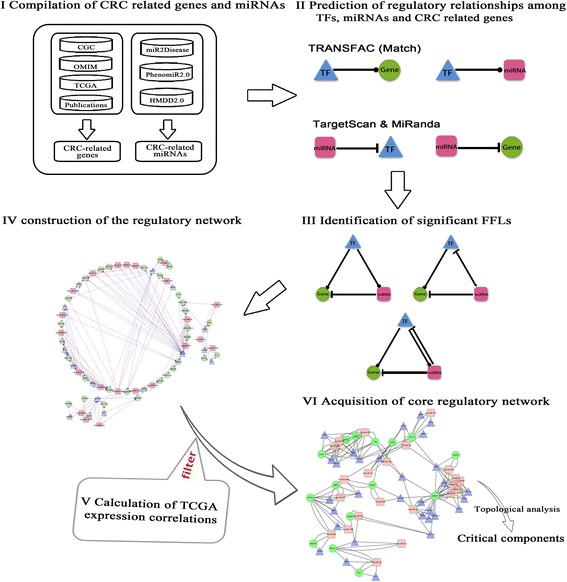
Process of miRNA-TF regulatory network construction and significant FFLs identification in colorectal cancer (CRC). This process contains six steps. 1) Data compilation. We extracted CRC-related genes, CRC-related microRNAs (miRNAs), and human transcription factors (TFs) from multiple databases. 2) Prediction of the regulatory relationships. The four regulatory relationships include TF-gene, TF-miRNA, miRNA-gene, miRNA-TF. 3) Feed-forward loop identification. Based on the regulatory relationships above, the significant 3-node feed-forward loops were identified. 4) CRC-specific miRNA-TF regulatory network construction and further analysis by merging the FFLs identified in step three. 5) TCGA expression correlation calculation. We calculated the expression correlations of each pair in the network, and removed the false positive pairs. 6) Acquisition of significant FFLs. We extracted the core subnetwork based on the significant pairs identified in step five. Furthermore, identification of critical miRNA and gene components were performed
