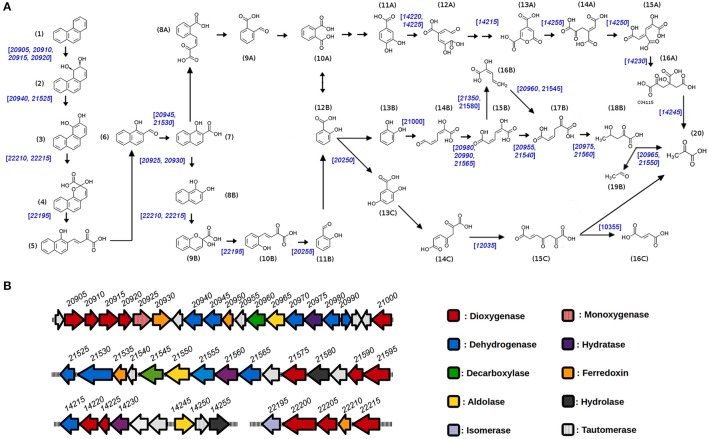
Figure 5.
Genetics of Phenanthrene degradation in S. xenophagum D43FB. (A) Predicted pathways of aerobic Phenanthrene degradation are shown, and genes mapped to D43FB genome with required enzymatic activies are shown in blue numbers. Chemical structures: (1) phenanthrene, (2) cis-3,4-Dihydrophenanthrene-3,4-diol, (3) phenanthrene 3,4-diol, (4) 2-hydroxy-2H-benzo[h]chromene-2-carboxylate, (5) cis-4-(1′-hydroxynaphth-2′-yl)-2-oxobut-3-enoate, (6) 1-hydroxy-2-naphthaldehyde, (7) 1-hydroxy-2-naphthoate, (8A) cis-2′-carboxybenzalpyruvate, (9A) 2-formylbenzoate, (10A) phthalate, (11A) protocatechuate, (12A) 4-carboxy-2-hydroxymuconate semialdehyde, (13A) pyrone-4,6-dicarboxylate, (14A) oxalomesaconate, (15A) 4-carboxy-2-hydroxy-cis,cis,-muconate, (16A) 4-hydroxy-4-carboxymethyl-2-oxoglutarate, (8B) naphthalene-1,2-diol, (9B) 2-hydroxychromene-2-carboxylate, (10B) trans-o-hydroxybenzylidenepyruvate, (11B) salicylaldehyde, (12B) salicylate, (13B) catechol, (14B) hydroxymuconate semialdehyde, (15B) hydroxymuconate, (16B) 2-hydroxy-pentadienoate, (17B) oxalocrotonate, (18B) 4-hydroxy-2-oxovalerate, (19B) acetaldehyde, (13C) gentisate, (14C) maleylpyruvate, (15C) fumarylpyruvate, (16C) fumarate, (20) pyruvate. (B) Organization of predicted Phenanthrene degradation genes in S. xenophagum D43FB. Principal operons encoding enzymatic functions involved in PAH degradation are shown, with predicted function and gene numbers.