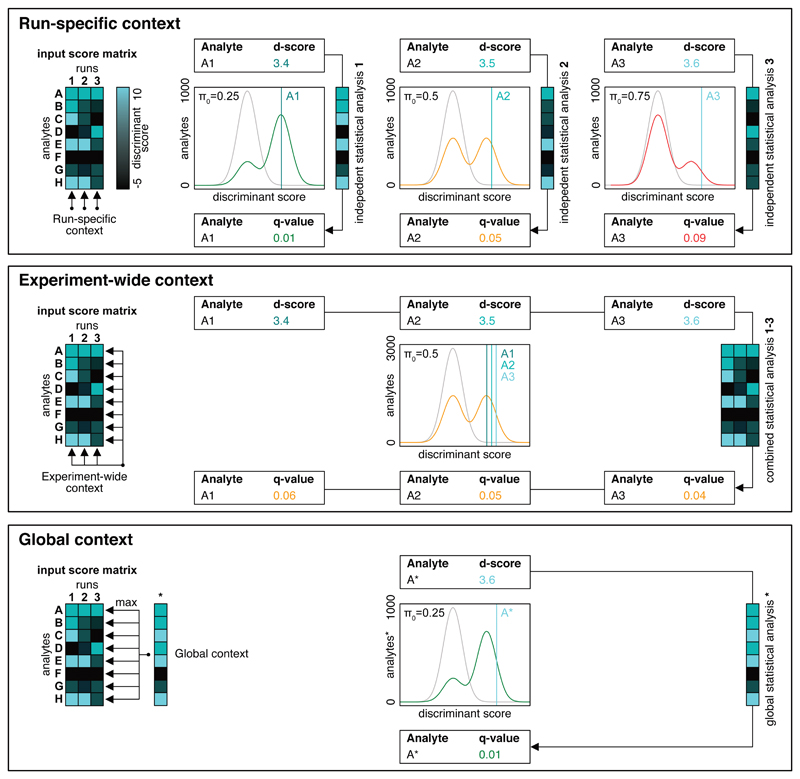
Figure 2. Schematic illustration of the different context-dependent error-estimation strategies.
The run-specific context conducts separate q-value estimation for each sample. This method results in run-specific q-values which can represent different peak group qualities between runs with varying π0. This means that if the same peptide is queried in two samples using the same parameters, run 1 with a low π0 and run 2 with a high π0, and the scored peak groups have a similar discriminant score (d-score), they might get a low q-value in run 1 and a high q-value in run 2. The experiment-wide context considers all runs of an experiment for error rate control. The resulting q-values can be compared in terms of peak group quality between runs but should not be considered outside the context of the whole experiment. The global context only considers the best scoring peak group per analyte across the entire experiment. This approach enables the total set of detectable peptides or inferred proteins to be determined within the experiment. The global set of proteins can optionally be used as a constraint for the experiment-wide context to obtain the number of detected analytes in single runs.