Figure 1. shRNA screening identifies Prmt1 as an essential gene for tumor-derived p53-deficient mOS cells.
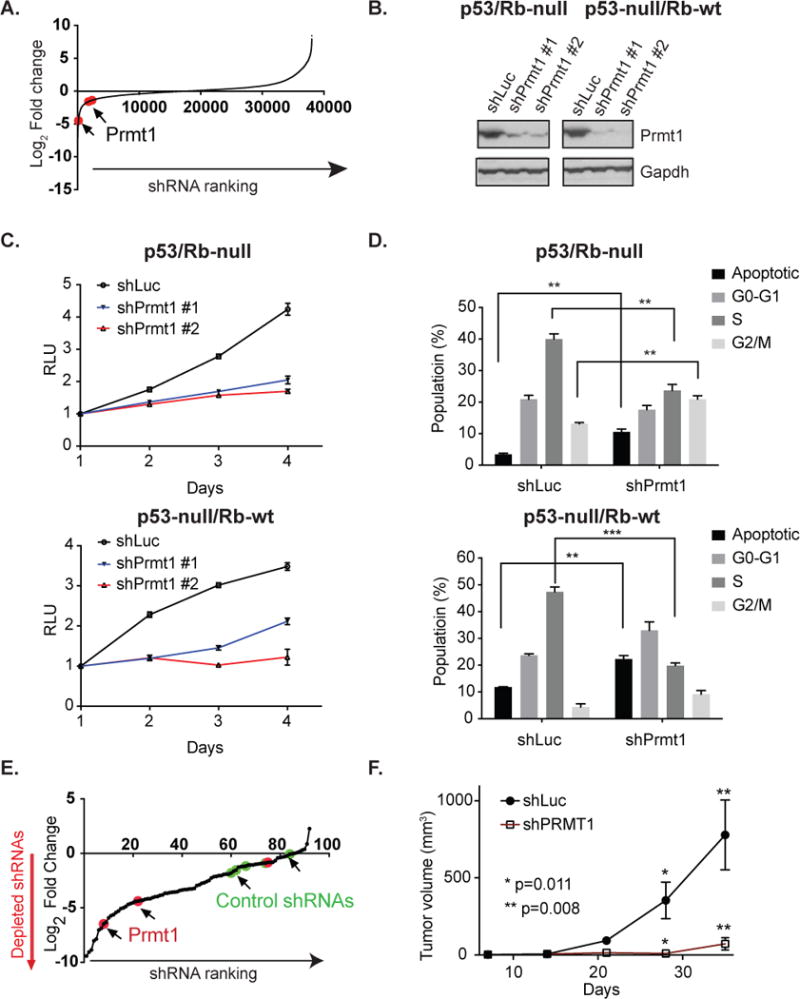
(A) Log2 fold change in shRNA abundance for p53/Rb-null mOS cell line at the end of the genome-scale in vitro shRNA screen relative to the initiatial reference pool. Prmt1-targeting shRNAs are highlighted in red. (B) Western blot analysis of Prmt1 expression in control (shLuc) and Prmt1 knockdown p53/Rb-null and p53-null/Rb-wt mOS cells. (C) Proliferation of p53/Rb-null and p53-null/Rb-wt mOS cell lines infected with non-targeting shRNA (shLuc) and Prmt1-targeting shRNAs. (D) Cell cycle analysis of p53/Rb-null and p53-null/Rb-wt mOS cells infected with control and Prmt1-targeting shRNA. The mean and standard deviation of triplicate samples are shown and t-tests were performed to determine the statistical significance between samples. ** p<0.01. *** p<0.001. (E) Log2 fold change in shRNA abundance for mOS xenografts relative to the initiation reference pool. shRNAs targeting Prmt1 are highlighted in red, while non-targeting control shRNAs are highlighted in green. (F) Growth of p53/Rb-null xenografts established using control shRNA (n=8) and Prmt1-targeting shRNA (n=8) infected mOS cells. The data are represented as mean + s.e.m. P values for the last time points are shown.
