Fig. 2.
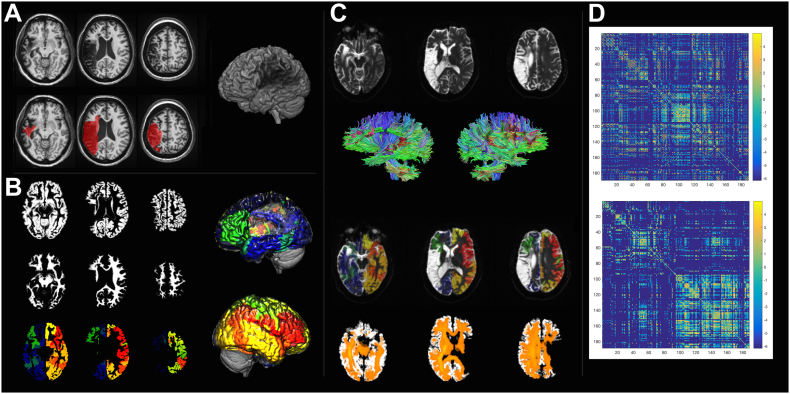
The methodological steps involved in the calculation of the connectome share similarities with VLSM. First, the necrotic/gliotic image is defined on T1 or T2 weighted images as shown in Panel A. Again, here we see a 3D render of an individual patient's brain with a lesion. Subsequently, an iterative segmentation and cost-function normalization approach is employed to define probabilistic maps of gray (Panel B, top row) and white matter (Panel B, middle row). The transformation matrix between T1 to MNI space is used to transfer an anatomical atlas to T1-weighted space and segment the probabilistic gray matter into regions of interest (Panel C, bottom row). Panel C also shows the 3D renders of segmentation into regions of interest (left and right lateral views with different colors for different regions). Tractography is performed in diffusion space, so the white matter mask and the segmented gray matter maps are transferred to B0 space (Panel C) and tractography is used to assess the number of streamlines linking each possible pairs of regions. Care is taken to ensure that tractography is performed being guided by the white matter probabilistic map, excluding the lesion site. The bottom row of Panel C shows a fiber density image in orange. Finally, a 2D matrix is generated where each entry represents the connection weight between the region in the row and column. The top matrix in Panel D shows the connectome, which is then arranged anatomically (Panel D, bottom matrix) to demonstrate the difference in the number of fibers in the left hemisphere (left upper matrix quadrant) versus the right hemisphere (right lower matrix quadrant).