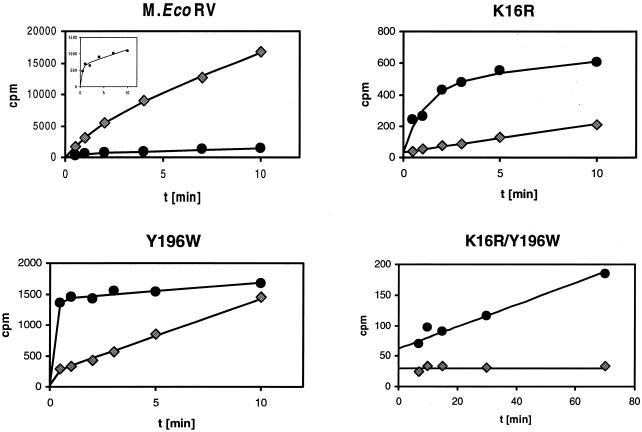
Figure 4.
Time courses of methylation of the adenine (RV_ME, diamonds) and the cytosine substrates (RV_C/A, circles) by M.EcoRV and three M.EcoRV variants. In these experiments, 0.5 µM oligonucleotide was incubated with 0.9 µM M.EcoRV or M.EcoRV mutant in methylation buffer containing 0.76 µM [methyl-3H]AdoMet. The insert in the panel showing the wild-type data displays an enlarged view on the reaction progress curve obtained with the cytosine substrate. All data points are averages of two to four individual measurements with errors always <10%. The lines show fits of the data points to equation 1 or linear regressions. The initial rates of DNA methylation of the K16R variant are 1.9 × 10–3 h–1 (adenine substrate) and 4.2 × 10–2 h–1 (cytosine substrate). The rate of methylation of the cytosine substrate by the K16R/Y196W variant is 2.0 × 10–4 h–1, whereas wild-type M.EcoRV modifies the cytosine substrate with a rate of 3.4 × 10–3 h–1 under these conditions (Table 1).