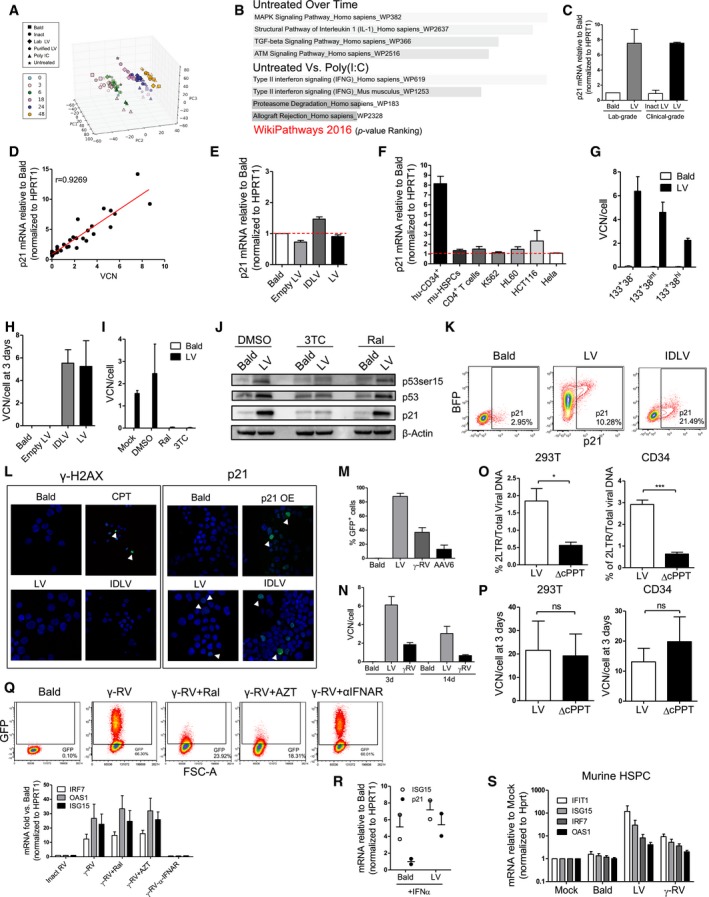
Figure EV1. Characterization of the transcriptional responses in HSPC.
-
APrincipal component analysis (PCA) plot. The samples from the RNA‐seq experiment are shown in the 3D plane spanned by their first three principal components. The first three principal components accounted for more than the 69% of explained variance among the data set.
-
BBar chart of the pathway enrichment analysis performed with WikiPathways database on the significant (P ≤ 0.05) differentially expressed genes over time in the untreated condition or between the Poly(I:C) and Mock conditions. The length of the bar represents the significance (P‐value Ranking) of that specific pathway.
-
Cp21 mRNA levels were measured after 48 h from the transduction of CB‐CD34+ with an MOI of 100 PGK‐GFP SIN LV (LV), MOI of 100 clinical‐grade LV (clinical LV), or to p24 equivalent of Env‐less (Bald) or inactivated clinical LV as controls (inactivated clinical LV) (mean ± SEM, n = 3).
-
Dp21 mRNA levels 48 h post‐transduction and vector copy numbers (VCN) 14 days after transduction were measured in CB‐CD34+ exposed to increasing MOI of PGK‐GFP SIN LV (LV).
-
Ep21 mRNA levels in CB‐CD34+ cells 5 days after transduction with an MOI 100 of LV, IDLV, or p24 equivalent of Bald or Empty LV as controls. Results are shown respect to Bald for each cell type set to value 1 (Red threshold) (mean ± SEM, n = 6).
-
Fp21 mRNA levels 48 days post‐transduction in different human and murine cell lines or primary cells exposed to PGK‐GFP SIN LV or p24 equivalent of Bald as control. p21 mRNA was measured 48 h after the transduction and normalized to HPRT1 for human cells or Hprt for Murine cells. Results are shown respect to Bald for each cell type set to value 1 (Red threshold) (mean ± SEM, n = 17 for hu‐CD34+, n = 8 for mu‐HSPC, n = 3 for CD4+ T cells and HCT116, n = 4 for HL60, n = 2 for K562 and Hela).
-
GVCN per cell were performed 14 days after exposure of sorted CB‐CD34+ to PGK‐GFP SIN LV (LV), at MOI 100 or p24 equivalents of Env‐less (Bald) as indicated (mean ± SEM, n = 5 for 133+38−, n = 8 for 133+38int and 133+38hi).
-
HCopy number per cell of total viral DNA 3 days after the transduction of human CB‐CD34+ exposed to an MOI of 100 PGK‐GFP SIN LV (LV), integrase‐defective LV (IDLV), p24 equivalent genome‐less (Empty LV), or Env‐less (Bald) (mean ± SEM, n = 4).
-
I, JVector copy number results and WB for p53 and p21 of CB‐CD34+ transduced with an MOI of 100 of PGK‐GFP SIN LV (LV), or Env‐less (Bald) as control. Transductions were performed in the presence of the integrase inhibitor Raltegravir (Ral), or the reverse‐transcriptase inhibitor Lamivudine (3TC). Control cells were transduced in DMSO or kept untreated (Mock). VCN and WB were performed 14 days and 48 h after transduction, respectively (mean ± SEM, n = 4, I; n = 1, J).
-
KRepresentative FACS plot of CB‐CD34+ cells 48 h after the transduction with PGK‐BFP LV, IDLV, or Bald as control and stained with anti‐p21 antibody.
-
LImmunofluorescence of CB‐CD34+ cells stained for γH2AX (left panel), 12 hours after the transduction with the indicated vector or camptothecin (CPT) exposure, and for p21 (right panel) 48 h after the transduction with PGK‐mCHERRY LV, IDLV, a p21 overexpressing LV (p21 OE) or Bald as a control (representative image of n = 2).
-
MPercentage of GFP positive CB‐CD34+ transduced with PGK‐GFP SIN LV at MOI 100 (LV), or exposed to p24 equivalent of Env‐less (Bald) vector as control, cells were also transduced with an MOI 100 of a PGK‐GFP SIN RV or to an MOI 10,000 of PGK‐GFP Adeno‐associated Vector (AAV6). GFP expression was assessed at FACS 5 days after transduction for the integrating vector, and 2 days post‐TD for the AAV6 (mean ± SEM, n = 9).
-
NVector copy number results of CB‐CD34+ transduced with PGK‐GFP SIN LV or PGK‐GFP SIN RV (RV) at MOI 100 or exposed to p24 equivalent of Env‐less (Bald) vector as control. Copy per cell of total viral DNA was assessed 3 days after transduction, while VCN on the integrated vectors were performed 14 days after transduction (mean ± SEM, n = 9).
-
O, P(O) Comparison of the nuclear import efficiency and (P) total viral DNA 3 days after transduction with LV or ΔcPPT. Nuclear import efficiency is reported as the percentage of 2LTR circles on the total reverse‐transcribed viral DNA. The same analysis was performed in 293T cells (upper panel) and in human CBCD34+ cells (MOI 100) (lower panel) (mean ± SEM, n = 4 for 293T, n = 12 for CD34+, Mann–Whitney test, *P = 0.0286, ***P < 0.0001, O), (mean ± SEM, n = 4 for 293T, n = 12 for CD34+, Mann–Whitney test, P).
-
QInterferon‐stimulated gene (ISG) expression in human CB‐CD34+ transduced with an MOI of 100 of PGK‐GFP SIN RV (γ‐RV), or heat‐inactivated RV (Inact RV) as control. Transductions were performed in the presence of the integrase inhibitor Raltegravir (Ral), reverse‐transcriptase inhibitor azidothymidine (AZT), or a neutralizing antibody against the human IFNα receptor (α‐IFNAR). FACS analysis of GFP expression was performed 2 days after the transduction (upper panel). ISGs (IRF7, OAS1, ISG15) were measured 48 h post‐transduction. Expression levels were normalized to HPRT1 and shown respect to Bald set to value of 1 (lower panel) (mean ± SEM, n = 6 for Inact RV, RV, RV + Ral, RV + AZT, n = 2 for RV + αIFNAR).
-
RISG15 and p21 mRNA levels in CB‐CD34+ cells exposed to 1,000 U/ml of human IFNα for 24 h and transduced for 48 h either with an MOI 100 of a PGK‐GFP LV or p24 equivalent of Bald as a control (mean ± SEM, n = 2).
-
SISG (IFIT1, IRF7, OAS1, and ISG15) induction in murine HSPC transduced with an MOI of 100 of PGK‐GFP SIN LV (LV), PGKGFP SIN RV (RV), or a p24 equivalent of Env‐less (Bald) 48 h post‐transduction. Cells were also left untreated as control in the mock condition. Expression levels were normalized to HPRT1 and shown respect to controls set to value of 1 (mean ± SEM, n = 3 for Mock, n = 8 for bald and LV, n = 4 for RV).
Source data are available online for this figure.
