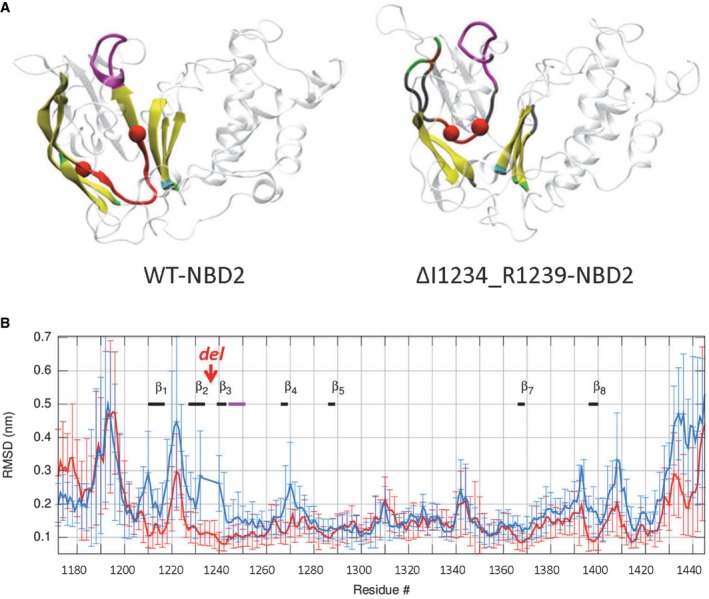
Representative snapshots of NBD2 in WT‐CFTR (left panel) and ΔI1234_R1239‐CFTR (right panel) systems following 30 ns of simulation. Each secondary structure element of the labeled β‐strands is represented by a unique color as follows: isolated‐bridge, brown; coil, gray; β‐strand, yellow; 3–10 helix, dark blue; α‐helix, cyan; turn, green; Walker A is in magenta, while the rest of NBD2 is in transparent representation. The amide N and carbonyl C atoms of residues 1,233 and 1,240, respectively, are shown as red spheres, and the backbone atoms between the same residue pair are shown in red cartoon representation.