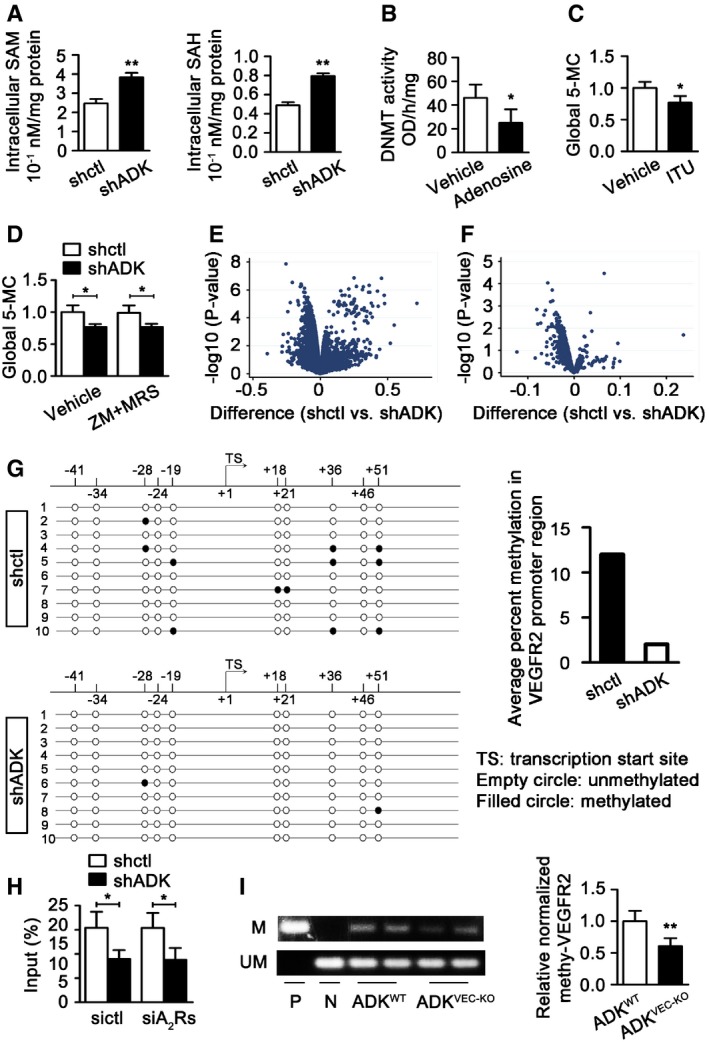
Quantification of intracellular SAM and SAH by HPLC in HUVECs. Results are from four independent experiments.
Quantification of relative DNMT activity in cell lysates of HUVECs incubated with or without 10 μM adenosine for 24 h. Results are from four independent experiments.
Quantification of 5‐mC levels in HUVECs treated with vehicle and ITU (10 μM) for 24 h. Results are from four independent experiments.
Quantification of 5‐mC in control and ADK KD HUVECs treated with or without both ZM 241385 and MRS 1754 at 5 μM for 24 h. Results are from four independent experiments.
Volcano plot showing standardized mean methylation difference of CpG sites among promoter regions comparing groups of shctl and shADK HUVECs (n = 3 independent cultures). x‐axis: mean methylation difference of CpG sites between groups of shctl and shADK HUVECs; y‐axis: −log10 of the P‐values.
Mean methylation difference of 1,261 CpG sites in the promoters of 109 angiogenic genes comparing groups of shctl and shADK endothelial cells. Number of replicates and axes as in panel (E).
(Left) DNA bisulfite analysis results for the region within 100 bp of the transcription start site of VEGFR2. (Right) Average methylation rate of CpG elements of VEGFR2 promoter region revealed by analysis of 10 randomly selected clones.
Methylated DNA immunoprecipitation results for VEGFR2 in ADK KD or Ctrl HUVECs transiently transfected with control or A2AR/A2BR siRNAs. Quantitative PCR was performed with purified total input DNA (input), and the DNA immunoprecipitated with 5‐mC antibody. Results are from four independent experiments.
Methylation‐specific PCR analysis of methylation status of VEGFR2 promoter of MAECs. P, positive control; N, negative control. Results are from four independent experiments.
Data information: For all bar graphs, data are the means ± SD, *
P <
0.05, and **
P <
0.01; unpaired, two‐tailed Student's
t‐test for (A–C) and (I); one‐way ANOVA with Tukey's
post‐hoc test for (D and H). The exact
P‐values are specified in
Appendix Table S1.
Source data are available online for this figure.