Abstract
Background
HOX antisense transcript RNA (HOTAIR) is a 2148 nt long, intergenic, non‐coding RNA molecule, which is reported to be highly expressed in many types of cancers. This meta‐analysis summarizes its expression in cancer.
Methods
We searched all eligible papers on the prognostic impact of HOTAIR in cancer from inception to 30 September 2015 in PubMed, CBMdisc, and the CNKI database. Only full texts were included. Revman 5.3 was used for meta‐analysis.
Results
A total of 11 studies of 1010 cases were included in the meta‐analysis. HOTAIR expression was higher in: cancer tissues in than adjacent or normal tissues (odds ratio [OR] 37.52, 95% confidence interval [CI] 18.94–74.31; P < 0.00001); in cancer tissues with lymph node metastasis than in those without lymph node metastasis (OR 3.37, 95% CI 2.36–4.82; P < 0.00001); and in histological grades II–III than in histological gradeI(OR 0.47, 95% CI 0.29–0.75; P = 0.002).
Conclusion
This study shows that HOTAIR may play an important role in cancer occurrence and development, but whether it is a marker of cancer diagnosis and reliable prognosis remains to be confirmed. More rigorous design and meticulous quality epidemiological studies are required.
Keywords: Cancer, HOTAIR, long non‐coding RNA, meta‐analysis
Introduction
Most human genomes are transcribed to RNA, but only 2% of RNA encodes functional proteins.1, 2 Long non‐coding RNA (lncRNA) more than 200 nt long is a transcription of RNA. It does not code for proteins, and most are located in the nucleus.3 lncRNAs have been described as transcriptional “noise,” existing to regulate gene expression levels.4 A class of small noncoding RNAs (ncRNAs) called microRNAs (miRNAs) have been implicated in the progression, prognosis, and therapy of malignant tumors.5lncRNA is also extensively involved in physiological and pathological processes, and plays an important role in the development of malignant tumors. HOX antisense transcript RNA (HOTAIR) is the first trans‐acting RNA to be found and is closely related to breast,6 liver,7 and colorectal cancers,8and other tumors. It may also represent a new diagnostic and prognostic molecular marker of early stage tumors. HOTAIR plays an important role in gene regulation by modifying chromatin structure.2, 9, 10GLOBOCAN reported that in 2012, there were 14 million new cases and 8.2 million patients died of cancer.11 Cancer has become the main cause of death worldwide. Five‐year survival rates for most cancers are still low. lncRNA recognizes the tumor and its molecular biology capabilities, which provides us with a new method to study the molecular biology and process of malignant tumors. HOTAIR has been associated with the occurrence and development of tumors; therefore, we used meta‐analysis to investigate the correlation between HOTAIR and cancer prognosis in this study.
Methods
Inclusion and exclusion criteria
Articles investigating the role of HOTAIR in the development of cancer with the following criteria were included in the study: (i) HOTAIR expression levels in primary cancerous tissues were measured, (ii) patients were grouped according to HOTAIR expression levels, (iii) complete clinical and pathological data were available, (iv) patients had not received chemotherapy or radiotherapy, and (v) sufficient data for the computation of odds ratios (OR) and corresponding 95% confidence intervals (CI) was available.
Repeated data or duplicated published literature, animal experiments, and case reports were excluded.
Document retrieval
We searched PubMed, CBMdisc, CNKI and other databases for English articles using the keywords: “HOTAIR,” “carcinoma,” “neoplasm,” and “prognosis.”
Literature screening and data extraction
Two reviewers separately screened the literature and ruled out any literature that obviously did not meet the inclusion criteria. When there were objections to a particular article, we consulted the article author and discussed acceptance or rejection after analysis.
Data extraction included: paper title, author, publication year, patient information, the number of patients studied, the follow‐up period, cancer and lymph node metastasis, histological grade, and histological differentiation grade.
Quality assessment of primary studies
Referencing Lichtenstein et al.’s case‐control study evaluation guidelines, two investigators independently performed quality assessment.12 The study selection is detailed in Figure 1.
Figure 1.
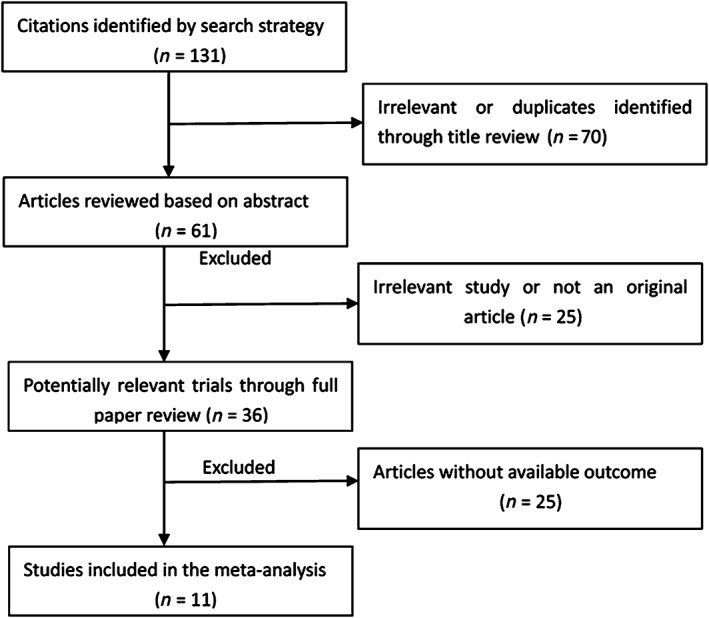
Flow diagram of the meta‐analysis.
Statistical analysis
We used RevMan 5.3 software to perform the meta‐analysis, counted the data using odds ratios (OR) and 95% confidence interval (CI) merger statistics, and 0.05 was considered statistically significant.13 The χ2 test was used to analyze statistical heterogeneity between studies, assuming a test level of α = 0.1. I2 was used for quantitative analysis of statistic heterogeneity. When P ≥ 0.1 and I2 ≤ 50%, no statistical heterogeneity was found between studies and a fixed‐effect model was used for meta‐analysis. When P < 0.1 and I2 > 50%, demonstrating high statistical heterogeneity between study results, or if the clinical heterogeneity was not apparent, a random effects model was used.
Results
Literature screening process
Thirty‐six articles were selected and after reading the title, abstract, and full text, we ultimately included 11 case‐control trials with a total of 1010 patients.
Basic characteristics and quality evaluation of included studies
The 11 selected studies included 160 cases of nasopharyngeal carcinoma, 171 cases of esophageal cancer, 119 cases of non‐small cell lung cancer, 87 cases of endometrial cancer, 132 cases of ovarian cancer, 110 cases of bladder cancer, 111 cases of cervical cancer, and 120 cases of colon cancer. Four studies reported HOTAIR expression in para‐carcinoma and cancer tissue or normal tissue,14, 15, 16, 17 eight reported HOTAIR expression with lymph nodes metastasis in carcinoma tissue,14, 15, 17, 18, 19, 20, 21, 22 and four reported HOTAIR expression in different pathologically graded carcinoma tissue.14, 22, 23, 24 A quality assessment and the basic characteristics of each study are shown in Table 1.
Table 1.
Basic characteristics and quality evaluation of included studies
| Study | Area | Tumor type | Gender (male/female) | Age (year) | N | Research index |
|---|---|---|---|---|---|---|
| Chen 2013 | China | Esophageal cancer | 50/28 | 61.9 ± 8.48 | 78 | † |
| He 2014 | China | Endometrial cancer | 87 | — | 87 | ‡ † |
| Kim 2015 | Korea | Cervical cancer | 111 | 50.6 ± 1.59 | 111 | † |
| Lv 2013 | China | Esophageal cancer | 56/37 | — | 93 | ‡ † |
| Nie 2013 | China | Nasopharyngeal carcinoma | 109/51 | 46 | 160 | ‡ † § |
| Takayuki 2013 | Japan | Non‐small cell lung cancer | 49/28 | — | 77 | † |
| Wu 2014 | China | Colon cancer | 64/56 | — | 120 | † |
| Liu 2013 | China | Non‐small cell lung cancer | 32/10 | — | 42 | ‡ |
| Qiu 2014 | China | Ovarian cancer | 64 | — | 64 | † § |
| Qiu 2015 | China | Ovarian cancer | 68 | — | 68 | § |
| Yan2014 | China | Bladder cancer | 80/30 | — | 110 | § |
HOTAIR expression with lymph node metastasis in carcinoma tissue.
HOX antisense transcript RNA (HOTAIR) expression in para‐carcinoma, cancer or normal tissues.
HOTAIR expression in different pathologically graded carcinoma tissue.
HOX antisense transcript RNA (HOTAIR) expression in cancer tissues and control
Four studies examined HOTAIR expression in cancer, cancer adjacent, and normal tissues. As there was no statistical heterogeneity among these studies (P = 0.25), the fixed effects model was adopted for meta‐analysis. HOTAIR expression in cancer tissues was higher than in the control group (OR 37.52, 95% CI 18.94–74.31; P < 0.000 01) (Fig 2). The shape of the funnel plot suggested that there was no publication bias (Fig 3).
Figure 2.
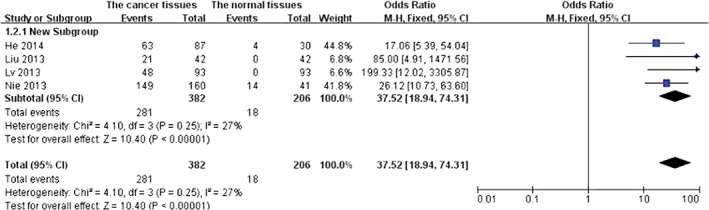
Analysis of HOX antisense transcript RNA expression in cancer versus control tissues. CI, confidence interval; M–H, Mantel–Haenszel.
Figure 3.
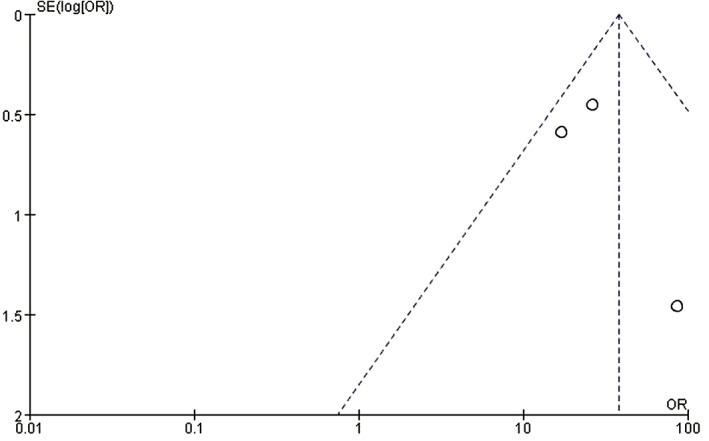
Funnel plot of publication bias. OR, odds ratio; SE, standard error.
HOTAIR expression in cancer tissues with and without lymph node metastasis
Eight studies examined HOTAIR expression in cancer tissues with and without lymph node metastasis.14, 15, 17, 18, 19, 20, 21, 22 There was statistical heterogeneity among these studies (P = 0.01); therefore, the random effects model was adopted for meta‐analysis. HOTAIR expression was higher in cancer tissues with lymph node metastasis than in those without (OR 3.37, 95% CI 2.36–4.82; P < 0.00001) (Fig 4). The shape of the funnel plot suggested that there was no publication bias (Fig 5).
Figure 4.
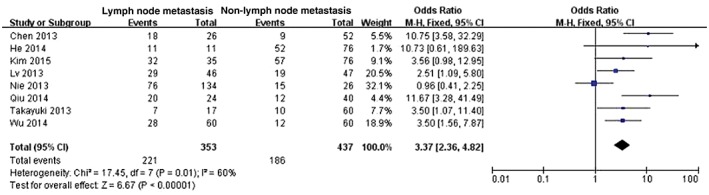
Analysis of HOX antisense transcript RNA expression is cancer tissues with and without lymph node metastasis. CI, confidence interval; M–H, Mantel–Haenszel.
Figure 5.
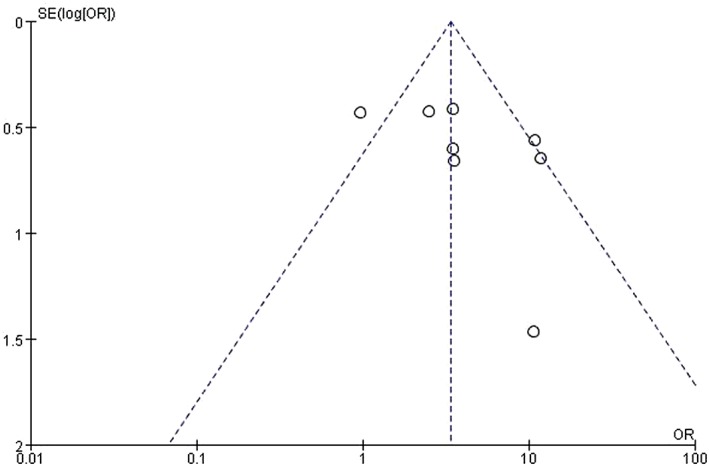
Funnel plot of publication bias. OR, odds ratio; SE, standard error.
HOTAIR expression in different histological classifications of cancer
Four studies examined HOTAIR expression in different histological classifications of cancer.14, 22, 23, 24As there was statistical heterogeneity among these studies (P = 0.02), the random effects model was adopted for meta‐analysis. HOTAIR expression was higher in cancer histological grades II–III (OR 0.47, 95% CI 0.29–0.75; P = 0.002) (Fig 6). The shape of the funnel plot suggested that there was no publication bias (Fig 7).
Figure 6.
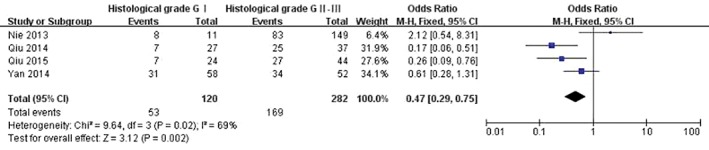
Analysis of HOX antisense transcript RNA expression between cancer histological gradesII–III. CI, confidence interval; M–H, Mantel–Haenszel.
Figure 7.
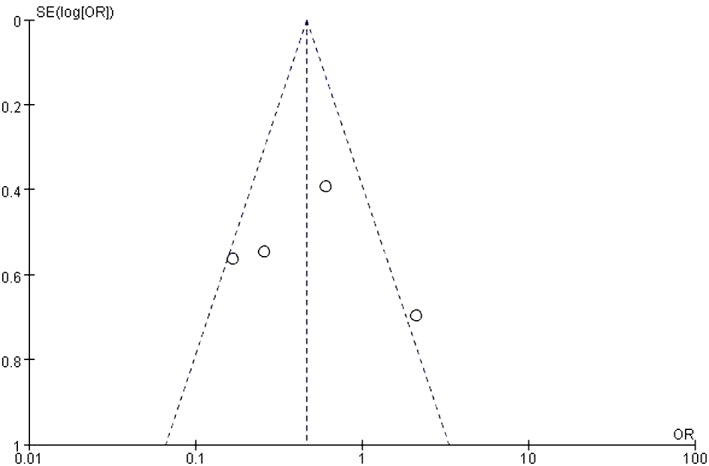
Funnel plot of publication bias. OR, odds ratio; SE, standard error.
Discussion
Eleven case‐control studies (1010 cases) examining the relationship between HOTAIR expression and cancer, lymph node metastasis, and histological grade were included in this meta‐analysis.14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24 HOTAIR expression in cancer tissues was higher than in normal tissues, and expression was correlated with lymph node metastasis and different histological grades.
Long intergenic non‐coding RNAs are deregulated in various human diseases, particularly cancer. lncRNAs in human disease might be correlated with their ability to impact cellular functions through different mechanisms.2, 25 Recently, studies have indicated that lncRNA HOTAIR plays a crucial role in the progression and metastasis of diverse cancers. HOTAIR is a 2158‐nucleotide lncRNA, a spliced and polyadenylated transcript located on chromosome 12q13.13. HOTAIR is highly conserved in primates and evolves faster than its neighboring HoxC genes. It negatively regulates transcription on another chromosome and is reported to reprogram chromatin organization and promote tumor progression. A number of studies have reported higher HOTAIR expression levels in paired primary cancerous tissues than in adjacent non‐cancerous tissues. HOTAIR overexpression is associated with high‐risk grade, metastasis, and poor overall survival in cancer patients. http://www.ncbi.nlm.nih.gov/pubmed/?term=Loewen%20G%5BAuthor%5D&cauthor=true&cauthor_uid=25491133 showed that HOTAIR represses gene expression by recruiting chromatin modifiers.26HOTAIR expression is elevated in lung cancer and correlates with metastasis and poor prognosis, promoting proliferation, survival, invasion, metastasis, and drug resistance in lung cancer cells. HOTAIR expression levels in colorectal cancer tissues are higher than those in corresponding non‐cancerous tissues and are associated with a poor prognosis.8 Gupta et al. showed that HOTAIR expression was associated with breast cancer metastasis.7 Kogo et al. speculated that HOTAIR expression was also associated with metastasis in colorectal cancer.8Using in vitro data, they showed that HOTAIR overexpression increased the invasiveness of colorectal cancer cells. These results indicate that HOTAIR might also play a role in promoting metastasis of colorectal cancer.
Using systematic review, we found that high HOTAIR expression is also associated with cancer occurrence, lymph node metastasis, and histological grade, with ORs of 8.15 (95% CI 4.61, 14.41), 1.83 (95% CI 1.06, 3.17), and 2.09 (95% CI 1.42, 3.08), respectively. Therefore, we believe that HOTAIR may be involved in the entire process of cancer occurrence and development.
It should be emphasized that there are several limitations to our study. First, although we conducted a relatively comprehensive search, we only reviewed published data and as such the lack of grey literature may have generated negative publication bias. Second, while stochastic study methods were described, the fact that no specific details of allocation concealment and how blinding was implemented were available may have introduced selection bias produced by human factors.
In summary, we found that HOTAIR expression in cancer tissues was higher than in adjacent and normal tissues, and was correlated with lymph node metastasis and different histological grades. HOTAIR may play an important role in the occurrence and development of cancer. However, the reliability of HOTAIR as a diagnostic and prognostic tool for cancer has not yet been confirmed. More rigorous, meticulous, and high quality design epidemiological studies are required.
Disclosure
No authors report any conflict of interest.
References
- 1. Bertone P, Stolc V, Royce TE et al. Global identification of human transcribed sequences with genome tiling arrays. Science 2004; 306: 2242–6. [DOI] [PubMed] [Google Scholar]
- 2. Carninci P, Kasukawa T, Katayama S et al. The transcriptional landscape of the mammalian genome. Science 2005; 309: 1559–63. [DOI] [PubMed] [Google Scholar]
- 3. Mattick JS, Makunin IV. Non‐coding RNA. Hum Mol Genet 2006; 15 (Spec No 1): R17–29. [DOI] [PubMed] [Google Scholar]
- 4. Ponjavic J, Ponting CP, Lunter G. Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Res 2007; 17: 556–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer 2006; 6: e857–66. [DOI] [PubMed] [Google Scholar]
- 6. Ishibashi M, Kogo R, Shibata K et al. Clinical significance of the expression of long non‐coding RNA HOTAIR in primary hepatocellular carcinoma. Oncol Rep 2013; 29: 946–50. [DOI] [PubMed] [Google Scholar]
- 7. Gupta RA, Shah N, Wang KC et al. Long non‐coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010; 464: 1071–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kogo R, Shimamura T, Mimori K et al. Long noncoding RNA HOTAIR regulates polycomb‐dependent chromatin modification and is associated with poor prognosis in colorectal cancers. (Published erratum appears in Cancer Res 2012; 72: 1039.) Cancer Res 2011; 71: 6320–6. [DOI] [PubMed] [Google Scholar]
- 9. Rinn JL, Kertesz M, Wang JK et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 2007; 129: 1311–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Khalil AM, Guttman M, Huarte M et al. Many human large intergenic noncoding RNAs associate with chromatin‐modifying complexes and affect gene expression. Proc Natl Acad Sci U S A 2009; 106: 11667–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. International Agency for Research on Cancer/World Health Organization . Globocan 2012: Estimated Cancer Incidence, Mortality and Prevalence Worldwide in 2012 [Cited 18 Jul 2014.] Available from URL: http://globocan.iarc.fr/Pages/fact_sheets_cancer.aspx. 2012.
- 12. Lichtenstein MJ, Mulrow CD, Elwood PC. Guidelines for reading case‐control studies. J Chronic Dis 1987; 40: 893–903. [DOI] [PubMed] [Google Scholar]
- 13. Zhou RY. Meta Analysis. Higher Education Press, Beijing: 2007. [Google Scholar]
- 14. Nie Y, Liu X, Qu S, Song E, Zou H, Gong C. Long non‐coding RNA HOTAIR is an independent prognostic marker for nasopharyngeal carcinoma progression and survival. Cancer Sci 2013; 104: 458–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lv XB, Lian GY, Wang HR, Song E, Yao H, Wang MH. Long noncoding RNA HOTAIR is a prognostic marker for esophageal squamous cell carcinoma progression and survival. PLoS ONE 2013; 8: e63516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liu XH, Liu ZL, Sun M, Liu J, Wang ZX, De W. The long non‐coding RNA HOTAIR indicates a poor prognosis and promotes metastasis in non‐small cell lung cancer. BMC Cancer 2013; 13: 464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. He X, Bao W, Li X et al. The long non‐coding RNA HOTAIR is upregulated in endometrial carcinoma and correlates with poor prognosis. Int J Mol Med 2014; 33: 325–32. [DOI] [PubMed] [Google Scholar]
- 18. Chen FJ, Sun M, Li SQ et al. Upregulation of the long non‐coding RNA HOTAIR promotes esophageal squamous cell carcinoma metastasis and poor prognosis. Mol Carcinog 2013; 52: 908–15. [DOI] [PubMed] [Google Scholar]
- 19. Nakagawa T, Endo H, Yokoyama M et al. Large noncoding RNA HOTAIR enhances aggressive biological behavior and is associated with short disease‐free survival in human non‐small cell lung cancer. Biochem Biophys Res Commun 2013; 436: 319–24. [DOI] [PubMed] [Google Scholar]
- 20. Wu ZH, Wang XL, Tang HM et al. Long non‐coding RNA HOTAIR is a powerful predictor of metastasis and poor prognosis and is associated with epithelial‐mesenchymal transition in colon cancer. Oncol Rep 2014; 32: 395–402. [DOI] [PubMed] [Google Scholar]
- 21. Kim HJ, Lee DW, Yim GW et al. Long non‐coding RNA HOTAIR is associated with human cervical cancer progression. Int J Oncol 2015; 46: 521–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Qiu JJ, Lin YY, Ye LC et al. Overexpression of long non‐coding RNA HOTAIR predicts poor patient prognosis and promotes tumor metastasis in epithelial ovarian cancer. Gynecol Oncol 2014; 134: 121–8. [DOI] [PubMed] [Google Scholar]
- 23. Yan TH, Lu SW, Huang YQ et al. Upregulation of the long noncoding RNA HOTAIR predicts recurrence in stage Ta/T1 bladder cancer. Tumour Biol 2014; 35: 10249–57. [DOI] [PubMed] [Google Scholar]
- 24. Qiu JJ, Wang Y, Ding JX, Jin HY, Yang G, Hua KQ. The long non‐coding RNA HOTAIR promotes the proliferation of serous ovarian cancer cells through the regulation of cell cycle arrest and apoptosis. Exp Cell Res 2015; 333: 238–48. [DOI] [PubMed] [Google Scholar]
- 25. Koch L. Disease genetics: Therapeutic targeting of a long non‐coding RNA. Nat Rev Genet 2015; 16: 2. [Google Scholar]
- 26. Loewen G, Jayawickramarajah J, Zhuo Y, Shan B. Functions of lncRNA HOTAIR in lung cancer. J Hematol Oncol 2014; 7: 90. [DOI] [PMC free article] [PubMed] [Google Scholar]


