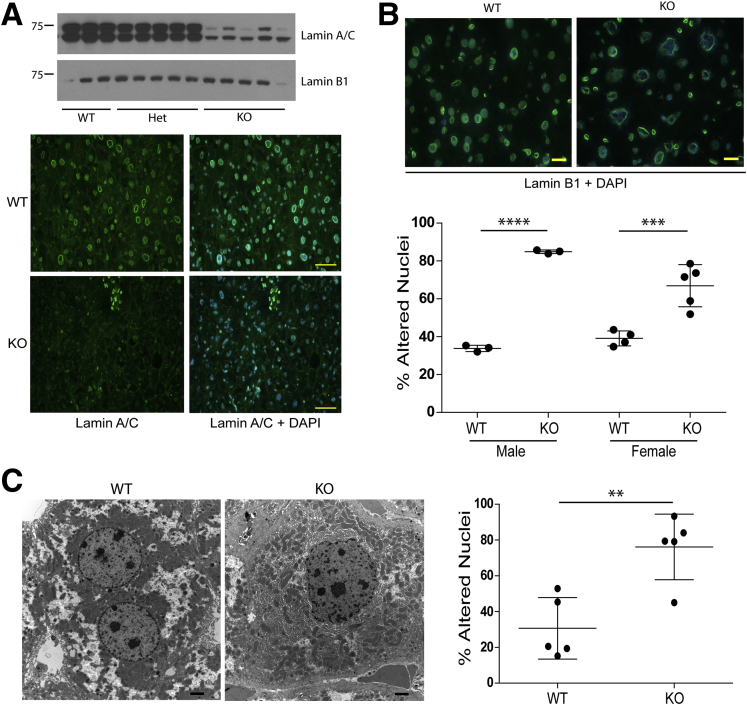
Figure 1.
Hepatic Lmna deficiency disrupts nuclear morphology. (A) Whole–cell extracts from Lmna WT, Het, and KO livers were analyzed by immunoblotting using antibodies to lamins A/C and B1. Each lane corresponds to a separate liver. Frozen sections from WT and KO livers were fixed in methanol and stained for lamin A/C followed by 4′,6-diamidino-2-phenylindole (DAPI). Scale bars: 50 μm. (B) Livers of 3 WT and 3 KO males (19–25 weeks of age) and 4 WT and 5 KO females (26–28 weeks of age) were cryosectioned and stained for Lamin B1 and DAPI. Normal-appearing and altered nuclei were counted in a blinded fashion from 5 fields per liver. Error bars represent SD, and statistical significance was determined by 1-way analysis of variance followed by the Tukey post hoc test. ****P < .0001 and ***P = .0005 for WT vs KO males and females, respectively. Scale bars: 20 μm for the representative sections shown. (C) Livers of 5 WT and 5 KO male mice (age, 22–24 wk) were examined by electron microscopy. Normal-appearing and altered nuclei (as defined by the presence of blebs and deformation) were counted in a blinded fashion from an average of 16 fields/liver. Error bars represent SD, and statistical significance (**P < .01) was determined using an unpaired t test (2-tailed). WT nuclei, n = 142; KO nuclei, n = 278. Scale bars: 2 μm for the representative sections shown.