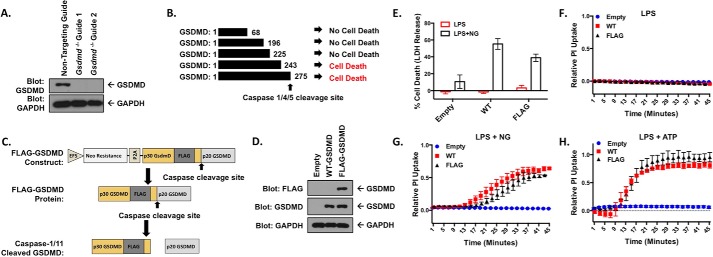
Figure 1.
Knock-out and reconstitution of gasdermin D in macrophages. A, CRISPR-Cas9 KO of Gsdmd in iBMDM cells. Cells transduced with each CRISPR guide were cloned out, individually verified for loss of GSDMD, and combined to form clonal pools of each guide. B, fragments of p30 GSDMD able to cause cell death identified by Shi et al. (9). C, design of the internal FLAG tag GSDMD construct. GSDMD is separated from the neomycin resistance marker by the P2A self-cleaving peptide, both under the control of an EFS promoter. The lentiviral expression construct allows for stable expression in multiple cell types. D, reconstitution of WT- and FLAG-GSDMD in Gsdmd−/− iBMDM cells. E, LDH release assay in reconstituted iBMDM cell lines primed with 200 ng/ml LPS and unstimulated or stimulated with 10 μm nigericin. F–H, PI uptake assay in WT- and FLAG- GSDMD iBMDM cells with LPS priming alone (F) and in combination with 10 μm nigericin (G) or 5 mm ATP (H). E represents means with S.E. of nine technical replicates of three experiments. F and H represent the means ± S.E. of four technical replicates of two independent experiments. G represents the means ± S.E. of six technical replicates of three independent experiments.