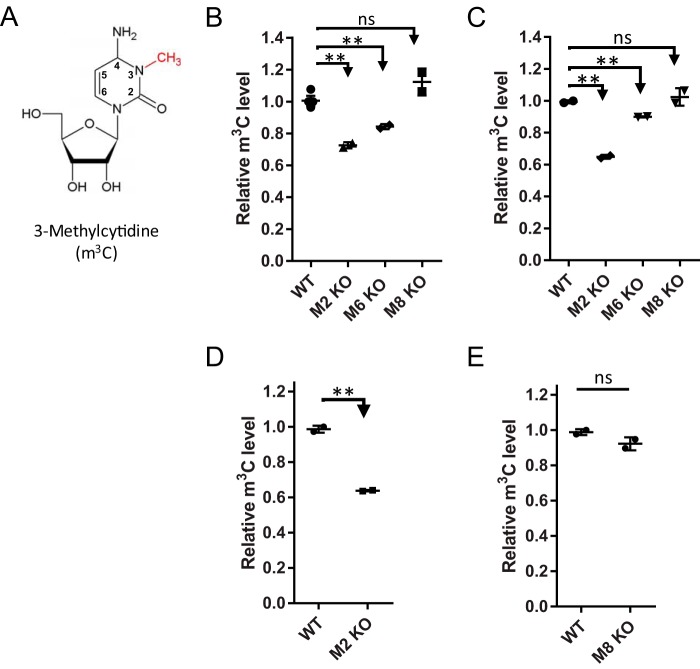
Figure 1.
Contribution of METTL2 and METTL6 to tRNA m3C in mouse and human cell lines. A, structure of m3C. Positions 2–4 are involved in Watson-Crick base pairing, with the N3-methylation serving as a block to polymerases. This polymerase-blocking phenomenon was exploited to map the location of methylations in tRNA. B and C, relative m3C levels in tRNA isolated liver and brain tissues, respectively. Samples from wild-type (WT), Mettl2 KO (M2 KO), Mettl6 mutant (M6 KO), and Mettl8 KO (M8 KO) mice were analyzed. D, relative m3C levels in HEK293T wild-type and METTL2 KO cell lines (loss of both METTL2A and -2B). E, relative m3C levels in HCT116 METTL8 wild-type and knock-out sample. m3C modification levels were normalized against levels of canonical cytidine. Data represent mean ± S.D. for at least three biological replicates, with asterisks denoting significant differences by Student's t test; **, p < 0.01. ns, not significant.