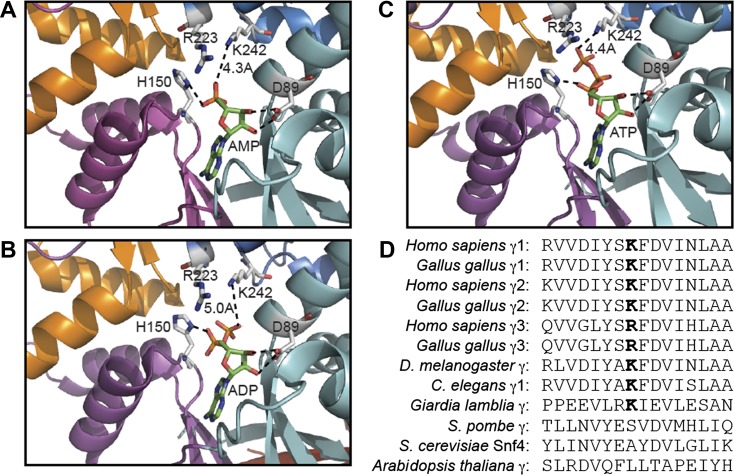
Fig. 4.
Structural analysis of the role of the equivalent K242 residue in rat γ1 and sequence conservation around K475 in AMPK isoforms. A−C: models of rat γ1 showing the relationship of D89, H150, R223, and K242 with bound AMP (A), ADP (B), and ATP (C) using RCSB Protein Data Bank ID 2V8Q, 2Y8L, and 2V9J, respectively (24, 30) (http://www.rcsb.org/pdb/home/home.do). Models were constructed using the MacPyMol molecular visualization system with CBS1 (cyan), CBS2 (magenta), CBS3 (blue), and CBS4 (orange) in “cartoon” representation and nucleotides and highlighted residues in stick representation (C atoms in grey in side chains and green in nucleotides, O atoms in blue, N atoms in red, and P atoms in orange). D: alignment of sequences around K475 in human γ2 and equivalent residues in other species and isoforms.