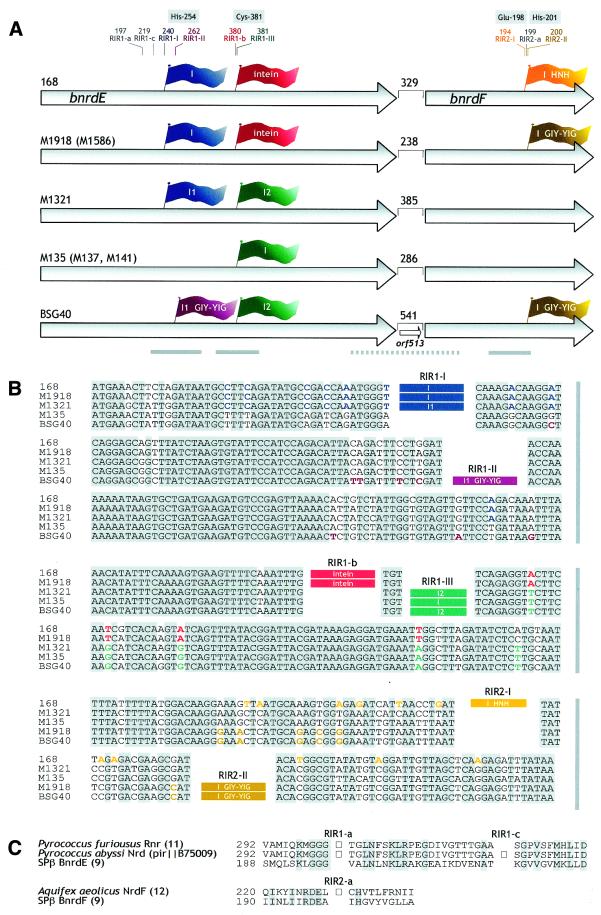
Figure 1.
(A) Schematic representation of IVS configurations in the bnrdE/bnrdF tandem of different Bacillus prophages. Arrows correspond to the bnrdE and bnrdF genes. The lengths of intergenic regions are given in nucleotides. A flag representing an IVS and its corresponding insertion site are in the same color. Occupied as well as unoccupied IVS insertion sites are represented by thin lines and annotated as follows: intron insertion sites, RIR1-I, RIR1-II, RIR1-III, RIR2-I and RIR2-II; insertion sites for intein DNA, RIR1-a, RIR1-b, RIR1-c and RIR2-a (13). Codons containing or preceding a given IVS insertion site refer to the sequence of hypothetical IVS– variants of the SPβ prophage bnrdE and bnrdF genes (9). Codons for some of the conserved and functionally important residues are boxed. I, intron. If two introns occupy the bnrdE gene, they are designated I1 and I2. GIY-YIG and HNH specify the intron homing endonuclease ORF present within the intron. Thick solid lines denote regions sequenced in all listed strains. Thick dotted lines represent segments sequenced in strains M1918, M1321, M135 and BSG40. Names of partly analyzed strains (see Results) are in parentheses. (B) Alignment of sequences surrounding IVS insertion sites in the bnrdE and bnrdF genes of Bacillus prophages. Conserved residues are shaded grey. IVSs more than 98% identical are specified by rectangles of the same color. Residues that are common to strains containing nearly identical IVSs, but absent from strains with an unrelated IVS, are colored accordingly. The intein DNA was deduced from the predicted intein sequence. (C) Alignment of sequences around archaeal intein insertion sites RIR1-a and RIR1-c, as well as the bacterial intein insertion site RIR2-a (13), with their unoccupied equivalents in the SPβ BnrdE and BnrdF proteins. Squares correspond to inteins.