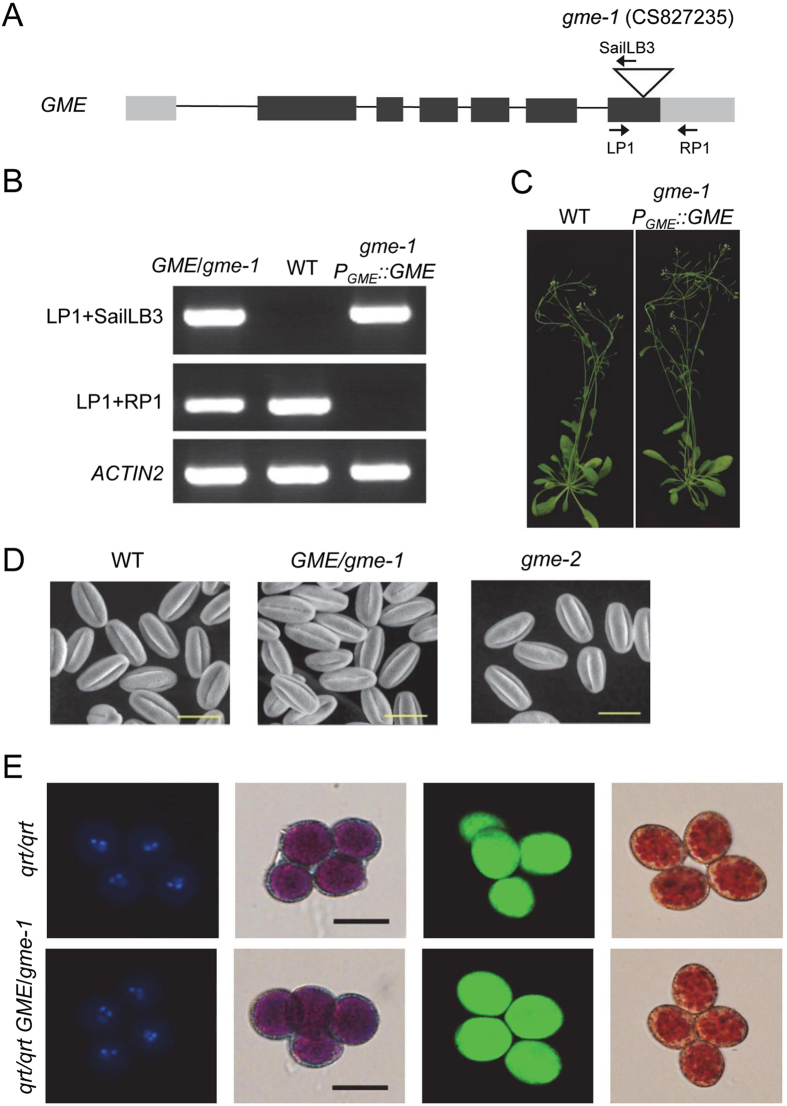
Figure 2.
Characterisation and genetic complementation of the gme-1 mutation and an analysis of tricellular pollen grains. (A) Schematic diagram showing the T-DNA insertion in GME and the core structure of the P GME::GME vector (GME under the control of the GME promoter). The grey rectangle, black rectangle and triangle represent the UTR, exon and T-DNA insertion site, respectively. The primers indicated by arrows were used to identify the genetic background of the gme-1 mutants in (B). (B) Genotyping of GME/gme-1, Col-0 wild type (WT), and transgenic P GME::GME in a gme-1 background. gme-1 P GME::GME was generated by transforming P GME::GME into GME/gme-1 heterozygous plants. The primer pairs LP1/SailLB3 and LP1/RP1, respectively, are specific for the gme-1 T-DNA insertion and WT GME. ACTIN2 PCR products were used as a control. (C) Six-week-old Col-0 WT and gme-1 P GME::GME plants. (D) Environmental scanning electron microscopy of pollen grains at floral stage 13 from WT, GME/gme-1 and gme-2 plants. Bars = 20 µm. (E) The gme-1 mutation does not affect male meiosis, mitosis, pollen viability or pollen vacuole development at the tricellular stage. The panels from left to right show tetrad pollen grains at the tricellular stage stained, respectively, with DAPI, Alexander’s stain, fluorescein diacetate/propidium iodide and neutral red. Bars = 20 µm.