Fig. 2.
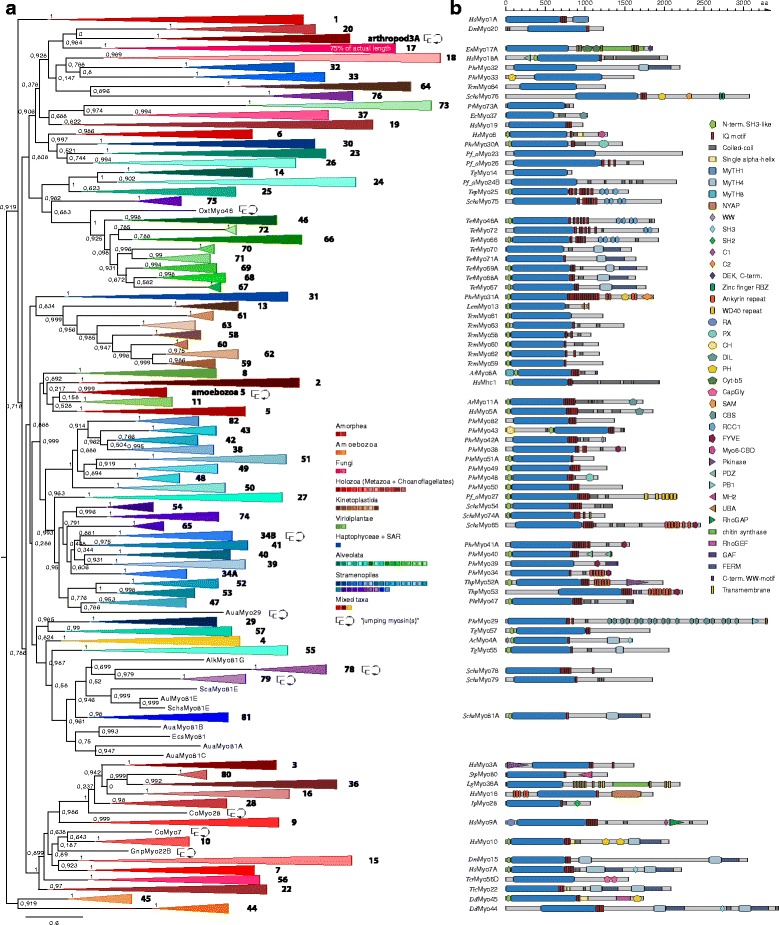
Myosin phylogeny and domain architecture. (a) Maximum-likelihood topology generated under the JTT + Γ model as implemented in FastTree. The tree is based on dataset6 applying a 90% sequence identity cut-off and removing orphans and some very divergent myosins resulting in 3104 myosin motor domain sequences (Additional file 1: Text). Although the entire myosin dataset is somewhat biased against metazoan and fungi species, the data used for the tree reconstruction is relatively balanced with not even two times more amorphean sequences as sequences from all other taxa. All branches containing only myosin members of a single class have been collapsed for better presentation. Some “jumping myosins”, for example the arthropod class-3A myosins (former class-21), do not group with the other members of their class in the tree of this dataset. Further trees of slightly different datasets are shown in Additional file 1: Figures S6 and S7. The scale bar represents the estimated number of amino acid substitutions per site. Myosin classes are coloured by class, not by taxa, but we used similar colours for taxon-specific classes as far as possible. (b) Scheme showing the domain architectures of selected members of the 79 myosin classes drawn to scale. The sequence name of a selected member of each class is given in the motor domain of the respective myosin. Regions not having assigned a defined domain do not necessarily indicate variable regions but rather missing domain definitions and might be highly conserved within the classes. Members have been chosen from well-known model species. However, they do not represent the full diversity of the domain architectures present in the respective classes. A key to all domain names and symbols but the motor domain is given on the right. The name of the representative myosin (species abbreviation + class and variant) is given in the myosin motor domain. Domain abbreviations are given in Additional file 1: Text. Species abbreviations are available in Additional file 2: Table S1
