Fig. 1.
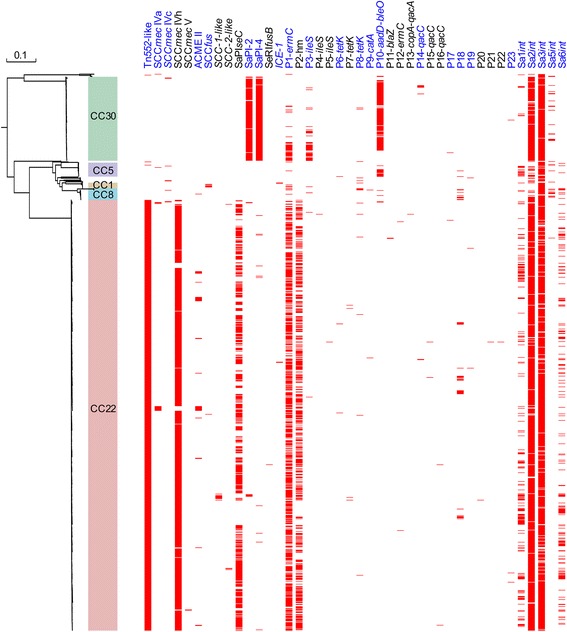
Phylogeny of 1193 S. aureus bloodstream isolates and distribution of analysed MGEs. The colour-coded column indicates clades representing the most common lineages: CC1, CC5, CC8, CC22 and CC30. All isolates were screened for carriage of MGEs detected in MRSA CC22. A rooted approximately maximum-likelihood phylogenetic tree was annotated with the distribution of all identified MGEs. Red horizontal lines indicate the presence of MGE. Names of MGEs detected in both CC22 and non-CC22 isolates are highlighted in blue. Putative plasmids were assigned a numeric ID plus the name of antimicrobial resistance gene (if applicable). For chromosomally integrated elements, ID names were assigned based on close homology to a previously described element as identified with Basic Local Alignment Search Tool (BLAST) based on sequence identity cut-off of 99%, or a numeric ID was assigned (name in italics)
