Abstract
Traditional in-house, laboratory-based medical imaging studies use hierarchical data structures (e.g., NFS file stores) or databases (e.g., COINS, XNAT) for storage and retrieval. The resulting performance from these approaches is, however, impeded by standard network switches since they can saturate network bandwidth during transfer from storage to processing nodes for even moderate-sized studies. To that end, a cloud-based “medical image processing-as-a-service” offers promise in utilizing the ecosystem of Apache Hadoop, which is a flexible framework providing distributed, scalable, fault tolerant storage and parallel computational modules, and HBase, which is a NoSQL database built atop Hadoop’s distributed file system. Despite this promise, HBase’s load distribution strategy of region split and merge is detrimental to the hierarchical organization of imaging data (e.g., project, subject, session, scan, slice).
This paper makes two contributions to address these concerns by describing key cloud engineering principles and technology enhancements we made to the Apache Hadoop ecosystem for medical imaging applications. First, we propose a row-key design for HBase, which is a necessary step that is driven by the hierarchical organization of imaging data. Second, we propose a novel data allocation policy within HBase to strongly enforce collocation of hierarchically related imaging data. The proposed enhancements accelerate data processing by minimizing network usage and localizing processing to machines where the data already exist. Moreover, our approach is amenable to the traditional scan, subject, and project-level analysis procedures, and is compatible with standard command line/scriptable image processing software. Experimental results for an illustrative sample of imaging data reveals that our new HBase policy results in a three-fold time improvement in conversion of classic DICOM to NiFTI file formats when compared with the default HBase region split policy, and nearly a six-fold improvement over a commonly available network file system (NFS) approach even for relatively small file sets. Moreover, file access latency is lower than network attached storage.
Keywords: Hadoop, HBase, Medical imaging, Grid computing
I. Introduction
Traditional grid computing approaches separate data storage from computation. To analyze data, each dataset must be copied from a storage archive, submitted to an execution node, processed, synthesized to a result, and results returned to a storage archive. This is the workflow traditionally adopted in processing medical imaging datasets. However, when imaging datasets become massive, the bottleneck associated with copying and ensuring consistency overwhelms the benefits of increasing the number of computational nodes. For example, consider the activity of converting Digital Imaging and Communications in Medicine (DICOM) files to NiFTI (a research file format); if converting a 50 MB volume takes 15 seconds, an ideal Gigabit network (≈ 100MB/s) saturates with slightly less than 30 simultaneous processes.
These challenges are further amplified considering the current trends where vast magnetic resonance imaging (MRI) and computed tomography (CT) databases are accumulating in radiology archives (at the rate of nearly 100 million examinations per year in the U.S.). However, we lack the image processing, statistical, and informatics tools for large-scale analysis and integration with other clinical information (e.g., genetics and medical histories). An efficient mechanism for query, retrieval, and analysis of all patient data (including imaging) would enable clinicians, statisticians, image scientists, and engineers to better design, optimize, and translate systems for personalized care into practice. Thus, a cloud-based service to address these needs holds promise.
It may however appear tempting to reuse existing cloud-based solutions for social networks and e-commerce, which provide a solution to this problem that is both simple and relatively inexpensive. These solutions combine the storage and execution nodes such that each task can be done with minimal copying of data. For example, the Apache Hadoop ecosystem [1], which provides Big Data processing capabilities, has been extensively used in these contexts.
Two reasons preclude such a naïve reuse. First, although such big data architectures have been applied in online commerce, social media, video streaming, high-energy physics, and proprietary corporate applications, these technologies have not been widely integrated with medical imaging data formats (e.g., DICOM) for medical image processing. Second, several approaches have followed the path of general machine learning literature and seek to implement algorithms specifically designed to take advantage of big data architecture [6], [11], [32], exploit the MapReduce framework to sift through datasets [25], or use distributed file systems [30], [36]. While such approaches have been effective for genetics studies [9], [36], they have not yet proven effective within current medical image computing workflows.
The fundamental reason for this shortcoming is that substantial resources have been invested in creating existing algorithms, software tools and pipelines, and hence there is a substantive (often prohibitive) cost associated with algorithm re-implementation and re-design specifically for big data medical imaging. Consequently, there is a need for new approaches that will not require algorithm re-design while still are able to exploit the potential of elastic, cloud-based frameworks, such as Apache Hadoop, that have shown promise in other application domains. However, as we show empirically, the default policies in the Apache Hadoop ecosystem are not effective in supporting big data medical imaging problems.
To address these problems, we present design principles and empirical validation for a new data model for use with cloud-based distributed storage and computation systems that provides practical access to distributed imaging archives, integrates with existing data workflows, and effectively functions with commodity hardware. Our approach makes specific improvements to the Apache Hadoop ecosystem, notably HBase, which is a NoSQL database built atop Hadoop’s distributed file system. Specifically, we make the following contributions:
A row-key design for Apache HBase: A hierarchical key structure is proposed as a necessary step to accommodate nested layers of priority for data-collocation.
New RegionSplit policy: A computationally efficient approach is proposed to optimally manage data collocation in the context of the hierarchical key structure.
Experimental results: The proposed innovations are evaluated in the context of a routine image analysis task (file format conversion) in a private research cloud comprising a typical Gigabit network with 12 nodes.
The performance of this new system is evaluated on small (7 GB) to moderate-sized (530 GB) test cases to characterize the overhead associated with this model and demonstrate tangible gains on widely available network and computational hardware. We believe that the proposed improvements to the Apache Hadoop ecosystem will greatly reduce the technical barriers to performing high-throughput image processing necessary to integrate imaging data into actionable metrics for personalized medicine. The novelty of the approach lies in our integrated solution for a novel application. The row-key design and linearizing most heavily used fields have been used in other contexts [21], [28], [31]. Yet, realizing new opportunities to apply existing techniques to a different realm (medical image processing), with its specific ontologies and patterns of data size/access, is essential to driving innovations. Our effort is a mix of research and experimental work to demonstrate applicability to medical imaging, which, to date, has not used a data-collocation computational model, and instead typically relies on monolithic data warehouses.
The rest of the paper is organized as follows: Section II describes our contributions; Section III describes our evaluation approach and presents experimental results; Section IV compares our work with related work; and finally Section V presents concluding remarks alluding to ongoing and future work, and discusses the broader applicability of our approach.
II. Enhancements to the Apache Eco-system
The task of processing medical images at scale requires a distributed image processing architecture that is aware of the underlying hierarchical imaging data and its meta-data. Our system is based upon the Hadoop framework, which was originally designed for file-system management and distributed processing [10], [14]. We combine Hadoop with Apache HBase, a NoSQL database which implements Google’s BigTable [8], [14]. The specific contribution of our work is a novel data storage mechanism that uses the hierarchical structure of imaging studies to collocate data with physical machines.
Before delving into our solution, we provide an overview of the problem domain we are working in, technologies we have used and the challenges faced.
A. DICOM and NiFTI Overview and Key Challenges
DICOM (Digital Imaging and Communications in Medicine) is the international standard for medical images and related information (ISO 12052). It defines the formats for medical images that can be exchanged with the data and quality necessary for clinical use (http://dicom.nema.org/). It has a hybrid structure that contains regular data (patient/clinical information), multimedia data (images, video). Data inside a DICOM file is formed as a group of attributes [27].
When a patient gets a Computed Tomography (CT) or Magnetic resonance imaging (MRI) scan, for example a patient’s brain image, a group of 2-dimensional DICOM images are generated slice by slice. A non-exhaustive set of medical imaging DICOM attributes for the slices include: project, subject, session and scan, where a project is a particular study, a subject is a participant within the study, a session is a single imaging event for the subject, and a scan is a single result from the event.
In order to study the entire brain, all 2-dimensional DICOM images should be collected together. Even though medical imaging data is stored as DICOM images, a substantial amount of medical image analysis software are NiFTI-aware (e.g. FSL (http://fsl.fmrib.ox.ac.uk/fsl/fslwiki/), AFNI (https://afni.nimh.nih.gov/afni/), SPM (http://www.fil.ion.ucl.ac.uk/spm/) and Freesurfer (http://freesurfer.net/)). NiFTI is a medical image data format, which is termed as a “short-term measure to facilitate inter-operation of functional MRI data analysis software package,” developed and founded by the NiFTI Data Format Working Group (http://nifti.nimh.nih.gov/).
Converting a large group of slices of DICOM images belonging to one patient into a small number of NiFTI format images (many-to-many relationship) is a significant step in any medical imaging study. Any processing of DICOM datasets will need to determine which CT/MRI scan that slice belongs to and using the Session attribute which records when the CT/MRI scan volume is carried out. However, finding a Session needs to first know the attribute Subject that it belongs to. Finally, the Project attribute collects all subjects together. Thus, for medical imaging applications involving DICOM, the following attributes are necessary: <monospace>project → subject → session → scan → slice</monospace>.
B. Apache HBase Overview and Key Challenges
HBase [26] uses the Hadoop Distributed File System (HDFS) to provide distributed and replicated access to data. We chose HBase since it can group data logically and physically based on row-key value. However, HDFS can only provide logical order. HBase also provides a flexible translation layer (region-split policy) for dealing with data collocalization statically or dynamically.
The key concepts from the HBase architecture are summarized in Table I. Briefly, HBase maintains tables, which have a row key that is commonly used as an index, and where data columns are stored with the row key. All data in HBase is “type free,” which are essentially in the format of a Byte Array. The table is sorted and stored based on the row key.
TABLE I.
HBase architecture key concepts summary
| Concept | Comment |
|---|---|
| Table/HTable | A collection of related data with a column-based format within HBase. |
| Region | HBase Tables are divided horizontally by row key range into “Regions.” A region contains all rows in the table between the region’s start key and end key. |
| Store | Data storage unit of HBase region. |
| HFile/Storefiles | The unit of Store, which is collocated with a Hadoop datanode and stored on HDFS. |
| memStore | When write data is uploaded to a HTable, it is initially saved in a cache as memStore. Once the cache size exceeds a pre-defined threshold, the memStore is flushed to HDFS and saved as HFile. |
| HMaster | HBase cluster master to monitor a RegionServer’s behavior for load balancing.Table operator. e.g., create,delete and update a table. |
| Regionserver | Serves read/write I/O of all regions in a cluster node. When Regionservers collocate with Hadoop datanode, it can achieve data locality. Subsequently, most reads are served by the RegionServer from the local disk and memory cache, and short circuit reads are enabled. |
| Rowkey | A unique identifier of a row record in table. |
| Column family | Columns in Apache HBase are grouped into column families. |
| Column identifier | The member in column family, also called as column qualifier. Multiple column identifiers can be used within one column family. |
HBase tables are divided into “regions” for distributed storage such that each region contains a continuous set of row keys from the overall table. The data in a region is physically collocated with an HDFS data node to provide data locality, which is performed by an operation called major compaction. When the region size grows above a pre-set physical size threshold, a “RegionSplitPolicy” takes effect and divides the region into smaller pieces. The newly created regions are automatically moved to different nodes for load balancing of the entire cluster. The row key and RegionSplitPolicy are integral to the performance and data retrieval of HBase and Hadoop.
Although HBase/HDFS is widely used in practice, multiple challenges manifest in the context of medical imaging applications. First, there is no standard for the default row key design. Intuitively, the data should be placed as sparse as possible and distributed evenly across various points of the regions in the table. Such a strategy can avoid data congestion in a single region, which otherwise could give rise to read/write hot-spots and lower the speed of data updates. Because row keys are sorted in HBase, using randomly generated keys when input as data to HBase would help leverage the data distribution in the table. As shown later, however, such an approach incurs performance penalties for medical imaging applications.
Second, since the original DICOM file name is a unique identifier called Global Unique identifier [17], if the task of interest is storing slice-wise DICOM data within HBase, then a naïve approach would be to use the DICOM GUID. Since the GUID is a hash of the data, it will not collocate data together and thus will saturate the network at the time of retrieving all DICOM images of a scan volume. Further, the standard RegionSplitPolicy will randomly assign files with hashed DICOM GUID file names as row keys to regions based on the key and a convenient split point based on region size, which may not be efficient as we show later.
C. Modified Row Key Design
To address the challenges outlined above, we propose a modified row key design for HBase based on the row key design requirements, which calls for preserving the structure of DICOM comprising the project, subject, session and scan. To maintain this structure, we propose using <monospace>< ProjectID > _ < SubjectID > _ < SessionID > _ < ScanID ></monospace> as the identifier with other optional characteristics, such as the “slice” appended to this identifier. This is how our collection of images are named in a hierarchical manner. For example, a row key looks like <monospace>Proj1_Subj2_Session3_Scan4_Slice5_example.dcm</monospace>, where “.dcm” is the filename extension for DICOM. Since HBase organizes data linearly based on row key, this new strategy will maintain data within a project that is split with a minimal, or just one more than the minimal number of splits across regions as possible, when used in conjunction with the default RegionSplitPolicy supported by HBase.
D. Modified RegionSplitPolicy for Medical Imaging
The default HBase policy that we evaluate and compare against is the <monospace>IncreasingToUpperBoundRegion-SplitPolicy</monospace> [2]. IncreasingToUpperBoundRegionSplitPolicy is based on a trigger point that dynamically updates itself from a specified minimum to a maximum (e.g., from 512 MB to 10 GB based on the total number of regions in a RegionServer). When a region split is triggered, the RegionServer first finds the largest files in the largest stores. The policy finds the first row key of the largest data block in each storefile. This key is called the “midkey” of a region and is decided based on region mid size. Thus, this split point can separate an existing associated imaging dataset into two regions without considering which row keys values lie in the split region. The newly created two regions will move through the whole cluster for storage balancing.
HBase also provides <monospace>KeyPrefixSplitKeyPolicy</monospace> (which inherits from IncreasingToUpperBoundRegionSplitPolicy) as one of the default split policy which is designed for grouping rows sharing a fixed pre-set length of keys [2]. However, it cannot dynamically group the subjects based on the order of project, subject, session etc based on highest available level (project,subject, session, scan) as we do in our modified approach. Moreover, it may cause a region to fail to split when all rows’ key have identical length of prefix (especially when prefix length is very short). This will make a region too large and impact the data balancing of the cluster. If the prefix length is very long, the effect of KeyPrefixSplitKeyPolicy would have no difference with IncreasingToUpperBoundRegionSplitPolicy. So we chose IncreasingToUpperBoundRegionSplitPolicy as the baseline comparison of HBase default split strategy.
To overcome these issues, we propose a novel RegionSplit-Policy, which has knowledge of the modified row key structure (see Section II-C) thereby maximizing data co-localization. It is critical to do it this way because typically users will select a cohort (set of subjects, sessions) for processing. The data under the same subjects or sessions are always processed together, not individually, e.g., in DICOM conversion the unit of processing is scan volumes. Thus, it is important to maximally collocate relevant image data under the same level for further group retrieval and processing/analysis, while reducing the data movement in MapReduce operations pertaining to the DICOM to NiFTI conversion, which is discussed in Section III-D1.
Our new RegionSplitPolicy also inherits from Increasing-ToUpperBoundRegionSplitPolicy but does the split differently. First it considers all row keys in a region. If multiple projects exist in the region, it splits the projects into separate regions. If the region is homogeneously a single project, it finds the highest available level (subject, session, scan) in the region on which it can split and balance the data between the new regions. Figure 1 compares the operation of the standard RegionSplitPolicy with our custom policy.
Fig. 1.
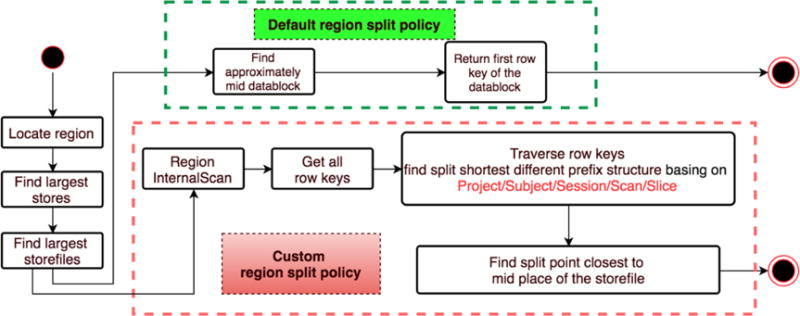
Comparison of the default RegionSplitPolicy (IncreasingToUpperBoundRegionSplitPolicy) and our custom RegionSplitPolicy. The standard policy splits the data within a region equally based on the data in the region. The custom policy considers the projects, subjects, sessions and scans in the region and makes a split to maximize data co-locality.
The challenge for our optimized RegionSplitPolicy is to find a split point based on all row keys of a Region. HBase provides a client API to retrieve data called scan (here, we refer to it as simple_scan). A user can customize the scan to define the range of row keys with which the column family and identifiers need to be retrieved. Users can also set customized filters to refine the query scan. A region has internal attributes that record the values of the start row key and end row key of the region. Since there are no attributes of records for any other row keys in a region except start/end row key, we need to use external ways to retrieve all row keys of a region.
In order to get all keys in a region, two existing approaches can be used in traditional HBase: (1) According to start/end key of region, a user specifies a column to scan. The scan is first executed on the entire table, finds the right RegionServer that hosts the region using Zookeeper quorum, and retrieves the row key; (2) Use HBase default RowKey filter to customize the scan. However, both approaches are slower compared to our approach described below.
As shown in Figure 1, we are capable of locating the largest storefiles. In this way, we can apply a more advanced HBase scan API (called “Region Internal scan”), which we have found to be 163 times faster than simple_scan on average in our tests to find all hierarchy row keys involved in the region. The Internal scan can directly operate on storefiles located on HDFS without starting a scan from the entire table. This gives us all the row keys of a split region. Next, the split point is selected according to the following conditions: (1) it ensures that the maximum related data is collocated in a hierarchy, and (2) once we have identified the level of structure which will be the potential point to split, we traverse the candidates and return the point that can most evenly balance the size of the two new regions in order to avoid the overhead of too many small regions emerging. Thus, if there are many projects of a region, we should split rows by <monospace>< ProjectID ></monospace>; if all row keys start with same project, and there is not only one subject, we should split rows by <monospace>< ProjectID > _ < SubjectID</monospace>, so on so forth.
The observed average run time range to determine one split point using our custom split policy is 28.22–58 ms, and 1.43–1.64 ms for the default HBase split policy with similar CPU usage (19.39% vs. 19.81%), which means the proposed split policy does not involve any substantial overhead when compared with the default one. The increased time in our policy is due to the need to retrieve and analysis all row keys of a region. Despite this one-time initial cost, as we show in our experimental results, the performance improvements are substantial.
E. Putting the Pieces Together
Figure 2 presents the overall structure of our modified Apache Hadoop ecosystem focusing particularly on the HBase modifications. HBase resides upon HDFS. Zookeeper monitors the health status of RegionServer. When users create a HBase table, they need to pinpoint the RegionSplitPolicy to HMaster, and the pre-set split policy is automatically triggered once when a Table region needs to be split. Our custom split policy is made the default split policy. The input DICOM is de-identified (for privacy preservation purposes) and is normalized to the hierarchy structure by a local row key generator before storing into HBase.
Fig. 2.
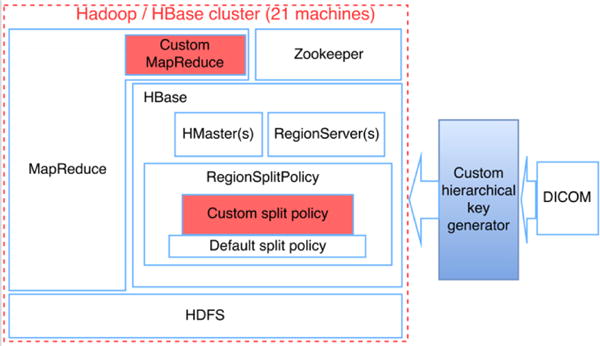
Overall Structure of Hadoop/HBase/Zookeeper Cluster with the Proposed Custom Row Key and Region Split Policy
F. Generality of the proposed approach
Our proposed approach has broader applicability than just DICOM2NiFTI. In recent work [4], we proposed a framework to evaluate the suitability of our approach for a larger set of medical imaging problems. Beyond that, multimedia processing always involves “large” data set compared with medical imaging data. For instance, video transcoding has been applied on Hadoop, each video is about 200 MB, and experiment data sizes are from 1 – 50 GB [20], [29]. Based on video record time and content, we can easily create a hierarchy category to name the video, and conduct group processing by omitting the reduce phase.
Gene data have many different styles with diverse attributes. Genes with similar expression patterns must be collocated for group analysis since genes that behave similarly might have a coordinated transcriptional response, possibly inferring a common function or regulatory elements [5]. Thus, genes data group/hierarchy storage, retrieval and analysis is applicable by our framework.
Another scenario where our work is applicable includes Satellite data/image processing on data about earth surface, weather, climate, geographic areas, vegetation, and natural phenomenon [16], which can be studied according to day-based, multiple-day-based, or seasonal-based [15]. As a result, time-oriented hierarchical structure can help group the data from the satellite for further processing. similarly, Internet of things collect data from various facilities like sensors. According to the sensors’ supervision area, a component hierarchy-based data collection can be implemented. For instance, high-speed train fault and repair prediction is applied before a train runs [34]. Analyzing mass historical data from a group of Electric Multiple Unit (EMU) of a train’s components has potential to be implemented in our framework.
III. Evaluational Methodology and Experimental Results
This section presents results of evaluating our Apache Hadoop/HBase modifications and comparing them with default strategies.
A. Testing Scenarios
To investigate the performance of our HBase modifications, we evaluated the standard DICOM to NiFTI file format conversion using three test scenarios using HBase and Hadoop, and one with Network Attached Storage (NAS) as follows.
Scenario: “Naïve HBase” – The project data was anonymized such that the original GUIDs were lost prior to this project and could not be recovered associated with data retrieval. True DICOM GUIDs are globally unique and contain both source (root stem) and random components. The MD5 tag mimics the random components from a single source vendor. Using MD5 hash key value meets the HBase original preferred key design for reducing hot-spot for table read/write. The DICOM files are distributed to all HBase regions, and we use an additional table to record the hierarchy structure of a scan dataset. We test using a random key, and MD5 hash of the data, as the key in HBase. With this comparison, we test the native capabilities of Hadoop and HBase without any of our proposed optimizations.
Scenario: “Custom Key/Default Split HBase” – This scenario evaluates the custom key grouping and ordering of the DICOM file logically and physically in HBase by our custom key value prefix introduced in Section II-C. When a HBase region exceeds a predefined size, we use the default split policy to split a region into two child regions without considering the key values of the split region. In this case, the files belonging to the same project, subject, session, scan are distributed into two different regions. The two regions may move to different cluster nodes, and the replication of both regions may also be placed on random Hadoop datanodes. When retrieving all files of a cohort (i.e., a set of scan volumes) for further processing, the MapReduce job dispatches computations to nodes that contain the datasets of interest. When no single node contains all requested data for a single job (either due to a large request or local storage scarcity), the minimal necessary data will be retrieved over network. So we test our proposed row key with the default RegionSplitPolicy.
Scenario: “Custom Key/Custom Split HBase” – Our custom RegionSplitPolicy has the capabilities to maximally collocate relevant data in the same group, with the order of project, subject, session and scans. We test our complete design with our proposed row key and custom RegionSplitPolicy and compare it with Custom Key/Default Split HBase to see how data retrieval matters in MapReduce. Theoretically, this approach involves less data collection and movement via the network than the other two HBase methods and makes processing faster.
Scenario: “Sun Grid Engine NAS” – Traditional grid computing approaches separate data storage from computation. As a comparative method, we use a traditional Sun grid engine (SGE) to distribute portable bash script (PBS) jobs to computational nodes accessing data from a Network Attached Storage (NAS) device.
B. Hardware
Twelve physical machines were used consisting of 108 cores of AMD Operon 4184 processors, 40 cores of Intel Xeon E5-2630 processors and 8 cores of Intel Xeon W3550 processors running Ubuntu 14.04.1 LTS (64 bit). At least 2 GB RAM was available per core. In total, 190 GB of storage was allocated to HDFS and a Gigabit network connected all of machines. The type for the local disks was Seagate ST32000542AS. Each machine was used as a Hadoop Datanode and HBase RegionServer for data locality. All machines were also configured using the Sun Grid Engine (Ubuntu Package: gridengine-* with a common master node). NAS was provided via CIFS using a Drobo 5N storage device (www.droboworks.com) with a 12 TB RAID6 array.
C. Data and Processing
To evaluate the test scenarios, 991,000 DICOM files from clinical CT scanners corresponding to 41 subjects and 812 scan volumes were retrieved in de-identified form under IRB approval from a study on traumatic brain injury. The processing system for each scan retrieves the data from storage (see test scenarios in Section III-A), applies a command line program and converts the DICOM files to NiFTI using dcm2nii (www.nitrc.org/projects/dcm2nii/). We performed tests with subsets of data with different datasets to assess the scalability of each proposed system and relative overhead (either fixed or scalable) versus processing load. To further test the system’s scalability and upper limit on throughput (i.e., number of datasets processed per minute), we incrementally increased the size of datasets 2, 4, … 10 times of the original 812 scan volumes. Thus, the average processing speed of one dataset from 812 to 8120 datasets scenario are same, and the impact of the fixed overhead will decrease as the number of datasets increase. Table II presents the dataset total file size for different number of datasets. Each dataset denotes the number of scans with average 126 DICOMs per scan.
TABLE II.
DICOM datasets size info
| Datasets | Total Scan size (GB) |
|---|---|
| 104 | 7.16 |
| 186 | 10.93 |
| 294 | 19.05 |
| 407 | 27.55 |
| 497 | 34.12 |
| 606 | 41 |
| 718 | 47.14 |
| 812 | 53.01 |
| 1624 | 106.02 |
| Datasets | Total Scan size (GB) |
|---|---|
| 2436 | 159.03 |
| 3248 | 212.04 |
| 4060 | 265.05 |
| 4872 | 318.06 |
| 5684 | 371.07 |
| 6496 | 424.08 |
| 7308 | 477.09 |
| 8120 | 530.1 |
D. Apache Hadoop/HBase Experimental Setup for DICOM2NiFTI
The Hadoop/HBase cluster is configured by Hadoop (2.7.1), HBase (1.1.2) and Zookeeper (3.4.6). HDFS uses default 3 replicas with rack awareness. In our experimental setup, the Sun grid engine does the balancing and makes sure that the jobs ran as soon as space was available within the specified node list when processing is executed on a traditional grid [13]. For Hadoop scenarios, MapReduce is a programming model and an associated implementation for processing large datasets in the Hadoop ecosystem [10]. YARN is used for resource (CPU/Memory) allocation and MapReduce job scheduling [33]. We use the default YARN capacity FIFO (First in First Out) scheduler, which aims at maximizing the throughput of cluster with capacity guarantees when the cluster is being shared with multi tenants.
The software tools to generate row keys from DICOM data were implemented in open source. The custom region split policy was implemented as a Hadoop extension class. All software is made available in open source at NITRC project Hadoop for data collocation (http://www.nitrc.org/projects/hadoop_2016/). Manual inspection of region stores was used to verify data collocation under multiple configurations of Hadoop Datanodes to ensure that the desired data collocation and region splits were occurring.
1) MapReduce Implementation of DICOM2NiFTI using HBase
The MapReduce model should complete two main tasks: data retrieval from HBase and data processing (DICOM to NIFTI conversion). The Map phase always tries to ensure that the computation tasks are dispatched where data resides, and those tasks are vividly called data-local maps. A compromise scenario is when data is not on the local node where the running map task is located, but at least the data are on the same rack, and those maps are rack-local maps. In the Reduce phase, the output, <monospace>< key, value ></monospace> pairs from Map phase are to be shuffled/sorted and sent to random cluster nodes.
If data retrieval is done in the Map phase while processing in the Reduce phase, then the computation and data can be on different nodes. A potential way to execute the processing is remote access (i.e. SSH) where data originally locates and applies processing. SSH limitations may occur, however, and block further connection, and as a result the processing cannot be fully completed. If both data retrieval and processing occurs in the Reduce phase, namely, each reduce task collects all row keys related with one scan volume, then downloads and collect DICOM files from HBase according to the keys, and finally executes the conversion in Reduce phase. In this strategy, network congestion will occur when the node that holds the Reduce task may not have all needed DICOM files. Since those DICOM files are aggregated in the same Region/node owing to our proposed custom split policy, so Reduce task has to retrieve all datasets through the network and will lead to congestion.
Thus, these approaches break our primary goal for data collocation with Hadoop and HBase with minimum data movement. As a result, our proposed Hadoop enhancements with data collocation in the context of the hierarchical key structure, data retrieval and processing occur in the Map phase and the Reduce phase is a no-op for our application.
In a traditional “word count” example, the input of MapReduce is a HDFS folder. The input folder is split into several pieces based on the files in the selected folder. Then each piece starts a map task with <monospace>< key, value ></monospace> pair, the input Map Key is file names and input Map value is file content. However, this approach is not practical in HBase. The HBase region has a corresponding folder on HDFS, and all data stores/hfiles in this region are placed in the region. When the region collocates to a Hadoop datanode to achieve data locality, all data store/hfiles are compacted to a giant file, which means that a traditional MapReduce like wordcount strategy cannot split an input HBase folder for further processing.
Figure 3 shows the modified work-flow. HBase provides a default API for running HBase-oriented MapReduce. The input of the MapReduce is a HBase-scan, which represents an interval of consecutive table row key values of a selected column. The HBase-scan is split based on relevant regions, and the input <monospace>< key, value ></monospace> pairs are values about row keys of a region and the content of the specified column. In short, if the input HBase-scan occurs across n regions, then only n map tasks are generated. The challenge for traditional HBase-oriented MapReduce for DICOM is there are usually more than one datasets of DICOM files under the same scan in a region. So we refined the above approach to specify the input of MapReduce to be a selected cohort of scan volumes, and the number of Map tasks is based on the number of scans.
Fig. 3.
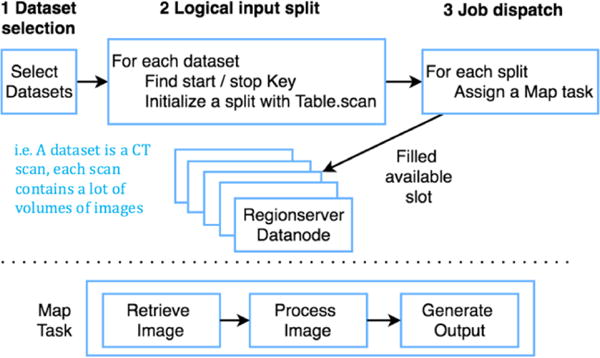
Custom HBase oriented MapReduce basing on input selected groups of scan volumes
DICOM with the same row-key prefix sticks together in order. Querying all DICOM images of a scan volume does not need to iterate over all input key values. Instead, we just need to define a search range (first/last row-key record of the selected cohort scan). Thus, we use an additional table to store the range of each scan volumes and do a one-time update once new images are uploaded to HBase. The Map Phase first retrieves the data from HBase and stores DICOM files to local node. Once done, it converts the DICOM files to NiFTI using dcm2nii as presented in Figure 3. For fair comparisons between Hadoop methods and approach on NAS, additional steps such as uploading the NiFTI result to HBase are not launched.
We did not include writing back to the original storage which is done typically in HBase to keep regions balanced and consistent across all cluster for each individual experiment. We did this because when writing back the result to HBase, the size of the region may change and may trigger a split, which takes time to localize a newly created region, particularly if it needs to be distributed to a different machine.
2) Guidelines used for Scaling Hadoop/HBase Cluster
Scalability is one of the most important properties for Cloud usage. We test and scale our clusters for studying intrinsic scalability performance. The following summarizes how we scaled the Hadoop/HBase cluster step by step.
For scaling down, RegionServer should first be gracefully stopped [2], and relationship of data collocation between Datanode and RegionServer are no longer exists. Then major compaction on the affected data from stopped RegionServer must be applied to collocate to the rest of the cluster [2]. When all data-locality is achieved again, decommissioning the Datanode and re-balancing of the cluster is performed. If decommission order is reversed, redundant replications are to be stored into HDFS which exponentially decreases the available size of the Hadoop cluster.
For scaling up, a new Hadoop Datanode must be commissioned first and then a new HBase RegionServer is added, followed by a major compaction to achieve data locality. If there is no Datanode, adding a new RegionServer can collocate to nothing, which makes reverse commissioning order no sense.
E. Results of Data Transfer Latency
The latency represents the data access latency for one dataset of slice images that belong to one project_subject_session_scan. First, we evaluated the latency in retrieving imaging data in each of the four scenarios. Table III shows average latency for all datasets. For naïve Hadoop, we retrieved data to a random node since the data were not collocated. For custom key/standard split, we retrieved the data to the machine which contained the first element in the scan. For custom key/custom split, we retrieved the data to the machine where the data were located entirely. For Grid Engine NAS, we retrieved the data from the NAS to a local machine serially (i.e., with one core in use).
TABLE III.
Latency results in seconds for each of the four test scenarios.
| Approach | Grid Engine NAS | Naive HBase | Custom key/Standard split HBase | Custom key/Custom split HBase |
|---|---|---|---|---|
| Latency(s) | 4.76 | 19.02 | 3.29 | 2.56 |
The naïve Hadoop strategy performed markedly worse than the other methods because it needs to open and close connections with multiple other machines in order to download the data, and the initialization and setup of each ZooKeeper connection involves overhead. Using the NAS with a single connection is relatively effective since the data are coming from one fixed location and there is low overhead in opening and closing connections. In comparing the default split policy to our proposed policy, we see an improvement in average performance. Any increase comes from the cases where scans are split between machines and thus data needs to be retrieved from other locations on the network which degrades latency.
F. Data Processing Throughput for DICOM to NiFTI Conversion
Each of the four scenarios executed a DICOM to NiFTI conversion as described in Section III-C. Figure 4A presents an analysis of throughput. The Grid Engine NAS performed the worst (fewest datasets per minute, longest run times) across all dataset sizes. In all scenarios, the NAS device saturated at 20 MB/s (approximately 18 datasets per minute) throughput despite the gigabit network access. This was likely due to numerous small files that are generated with “classic” DICOM scanning as direct read/write to the NAS device demonstrated substantively higher performance.
Fig. 4.
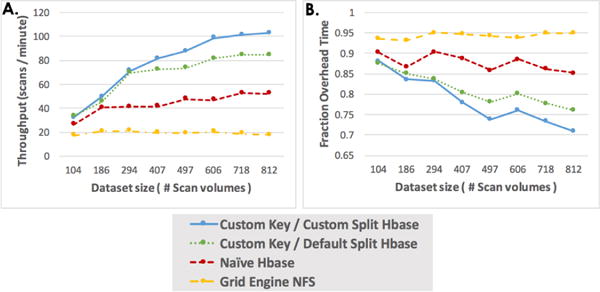
Throughput analysis for each of the test scenarios (104 – 812 datasets). (A) presents the number of datasets processed per minute by each of the scenarios as a function of the number of datasets selected for processing. (B) shows the fraction of time spent on overhead relative to the number of datasets.
The naïve HBase approach scaled better than the NAS approach with a throughput ranging from 31 MB/s (with 104 datasets) to 58 MB/s (with 718–812 datasets). The performance leveled off at 52 datasets/minute for a factor of almost threefold improvement over NAS. The custom key/default policy HBase approach performed even better with a throughput of 34 MB/s (with 104 datasets) to 94 MB/s (with 718–812 datasets). The custom key/custom policy HBase approach further increased throughput performance from 37 MB/s (with 104 datasets) to 114 MB/s (with 812 datasets).
The naïve method’s performance increases flatly because of uncertainty in the placement of data loading. It performs better than processing on the NAS device because not all data needs to be retrieved from the other node; some of the files are placed on same node with Map computation in most cases. On the other hand, the custom key custom policy HBase involves lesser data movement with better performance rather than the custom key/default policy HBase, both of whose processing are executed within most data-local map and a few rack-local map according to YARN allocation.
The root cause in reducing the processing time with the new policy is attributed to the following. Our approach (i.e., Custom key/Custom split HBase) groups the data based on the image’s hierarchical structure. When processing the data (i.e., DICOM to NiFTI conversion), a single task needs to load a subset of images that under the same structure (all image slices under the same project_subject_session_scan). Owing to the Hadoop MapReduce computation paradigm’s data locality feature, the job task can easily be dispatched to where the data is and hence the data retrieval is predominantly local so that it reduces the time for the computation to reach the data compared with SGE, which transfers data over the network.
1) Overhead Considerations with the Hadoop Framework
We define the overhead time as the difference between the actual wall clock time and the theoretical minimum time. The theoretical minimum time assumes all CPU slots are used except the last round of parallel jobs, and the average processing time of one dataset is empirically calculated by processing all pre-stored datasets on one machine without any data retrieval latency.
The computing grid had 156 cores available. Therefore, up to 156 jobs could run simultaneously in any of the test scenarios. With the three Hadoop scenarios, we have logs of both the time spent within each job on the compute node (including time to establish data connection, retrieve the data, and clean up the connection) and the actual wall clock time. For each of the Hadoop scenarios, we computed the average actual time spent executing the processing (including data retrieval), which ranged from 22 s to 35 s. For each of the data submission tasks, we can identify the minimum number of jobs that would need to run in serial by dividing the number of scan volumes by the number of cores. The fastest time that the Hadoop scheduler could run the jobs is the length of the serial queue times the job length, but in all cases the actual wall time exceeded this value.
The ratio of overhead time to total time is shown in Figure 4B. Fitting a linear analysis to each of the three scenarios shows that the SGE strategy had 95% overhead penalty due to data transfer latency. The naïve HBase strategy had a marginal penalty of 85% per additional dataset. The custom key/default split policy reduced the overhead penalty to 75% per additional dataset. Finally, the custom key/custom split policy resulted in 70% per additional dataset.
2) Upper Bound and Asymptotic Limits on Achievable Throughput
Figure 4A illustrates the processing on SGE, which saturates the Gigabit network. The HBase approach does not incur as much network congestion because most map tasks are data-local or rack-local. Thus, we were not able to observe any perceived network-imposed limitations even until 812 datasets. The upper limit on the throughput stems from other overheads in the framework, which we address in the paper. This is further verified in Section III-G. These overheads are either fixed or scale according to cluster size. Consequently, to reduce the impact of fixed overhead, we tested our system for more number of datasets.
To understand the asymptotic limits on the performance of our approach, consider Figure 5A which presents the result of processing scans per minute with more datasets according to Table II. We also provide the theoretical minimum time performance in Figure 5B. The data processing overhead is affected by the data retrieval and the fixed overhead stemming from MapReduce job initialization and zookeeper connection. The theoretical ideal solution fluctuates within the first 812 datasets due to differences in scan volumes. It then shows a flat performance because the datasets from 1,624 to 8,120 are duplicated from the first 812 datasets. We can see both Hadoop’s throughput gradually increased although the theoretical time oscillated at which point both Hadoop scenarios present a similar flat performance. These result trends empirically reveal the reduction of the impact of the fixed overhead.
Fig. 5.
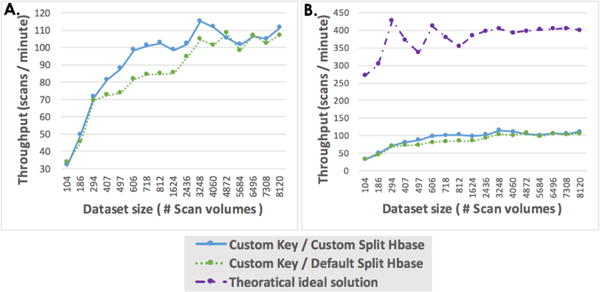
Throughput analysis for all test scenarios (104 – 8,120 datasets). (A) presents the number of datasets processed per minute by each of the scenarios as a function of the number of datasets selected for processing. (B) shows the relationship of throughput according to actual wall time and theoretical minimum ideal solution time.
Since the network is not a factor, we conclude that to obtain even higher throughput, we will need to scale the hardware by adding more cores since the number of cores is the limiting factor.
It is worth mentioning that to reduce the effort of reimplementing locally installed binary executables command-line problem our society mainly use (the program usually follows with system environment path), the proposed approach would temporally store the image byte array from HBase to a local place of the computation machine which arises overhead. For SGE scenario, since it uses NFS to attach the remoted NAS, the data retrieval is “like” to access local storage, so it does not need to temporally storage strategy as proposed Hadoop/HBase does. This paper aims to discuss and present empirical experiments result to proof the feasibility the framework.
G. Evaluating the Scalability of the Framework
We wanted to understand how does the scale of the cluster impact performance. Thus, we experimented by linearly decreasing the size of the cluster and observe if the performance decreased in similar manner. In our experiments, each machine acted as a Hadoop Datanode and HBase RegionServer for data locality as introduced in Section III-B. The order of decommisioning of Datanode and RegionServer is important when scaling the size of the cluster. Both the custom key with default and with custom split policy are compared on scaled cluster (5–10 Hadoop/HBase nodes). Decreasing the size of the cluster can linearly increase the total time with processing 5,684 datasets, which is presented in the trends of Figure 6A.
Fig. 6.
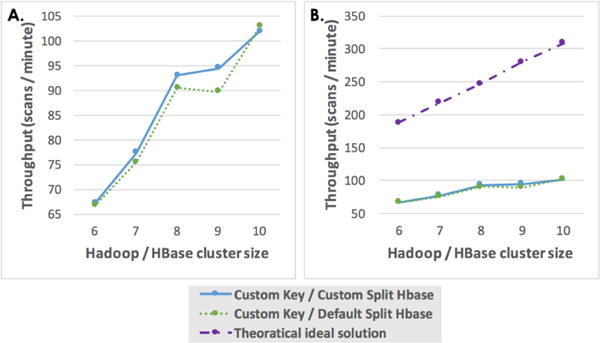
Throughput analysis for Hadoop scenarios (5,684 datasets) with different size of cluster (6 – 10 nodes, each node have 12 cores). (A) presents the comparison between two Hadoop scenarios with custom key. (B) shows the relationship between two Hadoop scenarios and theoretical ideal solution.
As mentioned before, DICOM to NiFTI conversion is the processing task we used in this research. One single task would use a dataset that has all the slices of images under the same dataset of project_subject_session_scan. The empirical results show that about 5% dataset of the scans would be split. Meanwhile, rack-local map cannot be ignored. We can conclude that the similar performance is due to the small proportion of split dataset in custom key/custom split strategy. If the dataset for a job is session-based rather than scan-based, the split ratio of dataset should be more distinctive, because custom key/custom split strategy groups images under same hierarchical structure better than default Hadoop/HBase strategy.
Based on the previous discussion, we can conclude that the Hadoop scenario performance is not limited by the network bandwidth but by the total available CPU cores and memory. While for SGE scenario, when scaling the cluster size would meet the meet deployed network bandwidth limitation. Thus, scaling up the size of the cluster can increase high performance computing capability for medical imaging processing in an affordable local/cloud-based commodity grid.
IV. Related Work
Recent trends indicate a substantial interest in adopting the MapReduce paradigm – and thereby the Apache Hadoop ecosystem – for medical image data processing. Several medical image processing studies have encountered one or more of the trio of computation, storage and network bandwidth bottlenecks, and have developed optimizations to overcome these encountered problems. This section compares our work with prior efforts in medical imaging and beyond.
A. Related Work involving Medical Imaging Applications
A recent study [25] illustrates how transitioning the medical image processing computations to the MapReduce paradigm and the Apache Hadoop framework pays rich dividends over traditional processing approaches, which often are sequential in nature. Our work differs from this prior work in that not only is our use case different – we focus on mapping DICOM images to NiFTI formats – but more importantly we demonstrate new optimization strategies for the Apache Hadoop ecosystem instead of simply leveraging the default strategies provided by Apache Hadoop, which is the case with most prior efforts. In fact the authors in this related work point out the need to identify opportunities for optimizations, which is precisely the intent of our presented research. Similarly, [19] demonstrates how the Apache Hadoop ecosystem can be used in medical imaging but do not report on any optimizations.
The work reported in [27] is synergistic to our work in that it focuses on the row- versus column-oriented storage issues for DICOM images. The authors highlight the pros and cons of row- versus column-oriented storage policies, and indicate how the complex structure of the DICOM images requires a hybrid mechanism for storage. Specifically, their approach stores frequently used attributes of a DICOM file into row-based layer/store, and optional/private attributes into a column-based store so that it will reduce null values. The motivation stems from the fact that if all DICOM attributes are stored into a row-based store, then a search or joining operation will unnecessarily involve numerous null values thereby adversely impacting efficiency.
The SYSEO project [7] also describes a hybrid row-column data store for DICOM images using similar criteria as in [27] to decide between row- versus column-based storage. Their work was motivated by the need to find alternatives to existing but prohibitively expensive solutions for medical image storage. Moreover, image annotation and query retrieval were additional dimensions that needed improvements in performance.
Our work do not treat DICOM file attributes as much depth as in [27], i.e., we need not to know the details of the attributes stored in a DICOM file when we store it to HBase; rather we simply store the entire DICOM file to HBase. For our DICOM to NiFTI processing, the processing operation can directly fetch the related attributes from DICOM files and convert them into NiFTI files. For other forms of medical imaging applications and data processing, such as image annotations, we may need to incorporate these hybrid storage mechanisms along with our optimizations, which forms our future work.
B. Related Work in other Application Domains
Several prior research efforts have proposed different performance optimizations to different elements of the Apache Hadoop ecosystem for domains beyond just medical image processing. The MHBase project [23] describes a distributed real-time query processing mechanism for meteorological data with the intent to provide safe storage and efficiency.
Recent work in Internet of Things (IOT) [24] proposes an optimization based on high update throughput and query efficient index framework including pre-splitting the HBase region for reducing the cost of data movement. Likewise, [22] addresses the problem of the HBase multidimensional data queries (upto four-dimension) in IOT with better response time. A recent work [35] demonstrates an optimized key-value pair schema for speeding up locating data and increase cache hit rate for biological transcriptomic data. The performance is compared with relational models in MySQL cluster and MongoDB. The authors in [18] present an optimized HBase table schema focusing on merging information to fit in combination with customer cluster and constructing an index factor scheme to improve the calculation of strategy analysis formulas.
In summary, the above-referenced prior efforts tend to focus on optimizing the table schema, row key design for data fast access, update and query. For our work, we not only provide an innovative row key hierarchical design, but also optimize the default RegionSplitPolicy which goes deep into the HBase architecture. Our goal is to maximally collocate relevant data on same node for further and faster group processing. Moreover, most prior works do not consider the cloud-based service aspect that we do.
V. Conclusions
Billions of magnetic resonance imaging (MRI) and computed tomography (CT) images on millions of individuals are currently stored in radiology archives [3]. These imaging data files are estimated to constitute one-third of the global storage demand [12], but are effectively trapped on storage media. The medical image computing community has heavily invested in algorithms, software, and expertise in technologies that assume that imaging volumes can be accessed in their entirety as needed (and without substantial penalty). Despite the promise of big data, traditional MapReduce and distributed machine learning frameworks (e.g., Apache Spark) are not often considered appropriate for “traditional”/“simple” parallelization.
In this paper we demonstrate that Apache Hadoop MapReduce can be used in place of a PBS cluster (e.g., Sun Grid Engine) and can be offered as a cloud-based service. Moreover, with our approach, even a naïve application of HBase results in improved performance over NAS using the same computation and network infrastructure.
We present a row key architecture that mirrors the commonly applied Project/Subject/Session/Scan hierarchy in medical imaging. This row key architecture improves throughput by 60% and reduces latency by 577% over the naïve approach. The custom split policy strongly enforces data collocation to further increase throughput by 21% and reduce latency by 29%. With these innovations, Apache Hadoop and HBase can readily be deployed as a service using commodity networks to address the needs of high throughput medical image computing.
Our experiments promote a general framework for medical imaging processing (e.g., structured data retrieval, access to locally installed binary executables/system resources, structured data storage) without comingling idiosyncratic issues related to image processing (e.g., parameter settings for local tissue models, smoothing kernels for denoising, options for image registration). DICOM2NiFTI is a routine first step in processing and often a bottleneck for quality control on large datasets. Hence, the application demonstrates the system’s correctness and scalability across a complex organization of files.
The system was implemented on a small, private data center, which includes the Sun Grid Engine. As the number of machines increases, NFS becomes nonviable with a single host, and distributed storage (e.g., GPFS) is commonly used on large clusters with 10+ Gbps networks. The proposed data and computation co-location solution is an alternative and could scale to well-more CPU-cores than beyond a GPFS solution on the same underlying network.
Finally, as implied by the trends in Figure-4, the benefits of distributing computation with storage increase with larger datasets. Exploration of the asymptotic performance limits is of great interest, but beyond the scope of this paper that illustrates meaningful gains on problems of widely applicable scale. The optimization of characterization of these approaches on heterogeneous grid is an area of great possibility. In particular, the Apache Hadoop YARN scheduler could be further optimized to exploit intrinsic relationships in medical imaging data.
The work presented in this paper is available in open source at www.nitrc.org/projects/hadoop_2016
Acknowledgments
This work was funded in part by NSF CAREER IIS 1452485 and US Ignite CNS 1531079. This work was conducted in part using the resources of the Advanced Computing Center for Research and Education at Vanderbilt University, Nashville, TN. This project was supported in part by the National Center for Research Resources, Grant UL1 RR024975-01, and is now at the National Center for Advancing Translational Sciences, Grant 2 UL1 TR000445-06. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of NSF.
References
- 1.Apache Hadoop Project Team. The Apache Hadoop Ecosystem. http://hadoop.apache.org/
- 2.Apache HBase Team. Apache hbase reference guide. Apache; Apr, 2016. version 2.0.0 edition. [Google Scholar]
- 3.AT&T. Technical Report AB-2246-01. AT&T; Jul, 2012. Medical imaging in the cloud. [Google Scholar]
- 4.Bao S, Weitendorf FD, Plassard AJ, Yuankai H, Gokhale A, Bennett LA. Theoretical and empirical comparison of big data image processing with apache hadoop and sun grid engine. SPIE Medical Imaging. 2017 doi: 10.1117/12.2254712. accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barrett T, Edgar R. [19] gene expression omnibus: Microarray data storage, submission, retrieval, and analysis. Methods in enzymology. 2006;411:352–369. doi: 10.1016/S0076-6879(06)11019-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bednarz T, Wang D, Arzhaeva Y, Lagerstrom R, Vallotton P, Burdett N, Khassapov A, Szul P, Chen S, Sun C, et al. Signal and Image Analysis for Biomedical and Life Sciences. Springer; 2015. Cloud Based Toolbox for Image Analysis, Processing and Reconstruction Tasks; pp. 191–205. [DOI] [PubMed] [Google Scholar]
- 7.Chabane Y, d’Orazio L, Gruenwald L, Mohamad B, Rey C. Medical Data Management in the SYSEO Project. ACM SIGMOD Record. 2013;42(3):48–53. [Google Scholar]
- 8.Chang F, Dean J, Ghemawat S, Hsieh WC, Wallach DA, Burrows M, Chandra T, Fikes A, Gruber RE. Bigtable: A Distributed Storage System for Structured Data. ACM Transactions on Computer Systems (TOCS) 2008;26(2):4. [Google Scholar]
- 9.Chen S, Bednarz T, Szul P, Wang D, Arzhaeva Y, Burdett N, Khassapov A, Zic J, Nepal S, Gurevey T, et al. Service-Oriented Computing–ICSOC 2013 Workshops. Springer; 2013. Galaxy+ Hadoop: Toward a Collaborative and Scalable Image Processing Toolbox in Cloud; pp. 339–351. [Google Scholar]
- 10.Dean J, Ghemawat S. MapReduce: Simplified Data Processing on Large Clusters. Communications of the ACM. 2008;51(1):107–113. [Google Scholar]
- 11.Freeman J, Vladimirov N, Kawashima T, Mu Y, Sofroniew NJ, Bennett DV, Rosen J, Yang CT, Looger LL, Ahrens MB. Mapping Brain Activity at Scale with Cluster Computing. Nature methods. 2014;11(9):941–950. doi: 10.1038/nmeth.3041. [DOI] [PubMed] [Google Scholar]
- 12.Frost and Sullivan. US Data Storage Management Markets for Healthcare. 2004 Nov; [Google Scholar]
- 13.Gentzsch W. Cluster Computing and the Grid, 2001 Proceedings First IEEE/ACM International Symposium on. IEEE; 2001. Sun grid engine: Towards creating a compute power grid; pp. 35–36. [Google Scholar]
- 14.Ghemawat S, Gobioff H, Leung ST. ACM SIGOPS operating systems review. Vol. 37. ACM; 2003. The Google File System; pp. 29–43. [Google Scholar]
- 15.Golpayegani N, Halem M. Cloud Computing, 2009 CLOUD’09 IEEE International Conference on. IEEE; 2009. Cloud computing for satellite data processing on high end compute clusters; pp. 88–92. [Google Scholar]
- 16.Gorgan D, Bacu V, Stefanut T, Rodila D, Mihon D. Intelligent Data Acquisition and Advanced Computing Systems: Technology and Applications, 2009 IDAACS 2009 IEEE International Workshop on. IEEE; 2009. Grid based satellite image processing platform for earth observation application development; pp. 247–252. [Google Scholar]
- 17.Hernández V, et al. Challenges and Opportunities of Healthgrids: Proceedings of Healthgrid 2006. Vol. 120. IOS Press; 2006. Bridging clinical information systems and grid middleware: a medical data manager; p. 14. [PubMed] [Google Scholar]
- 18.Hong S, Cho M, Shin S, Seon C, Song S, et al. 2016 8th International Conference on Knowledge and Smart Technology (KST) IEEE; 2016. Optimizing hbase table scheme for marketing strategy suggestion; pp. 313–316. [Google Scholar]
- 19.Jai-Andaloussi S, Elabdouli A, Chaffai A, Madrane N, Sekkaki A. 20th International Conference on Telecommunications (ICT), 2013. IEEE; 2013. Medical Content-based Image Retrieval by using the Hadoop Framework; pp. 1–5. [Google Scholar]
- 20.Kim M, Cui Y, Han S, Lee H. Towards efficient design and implementation of a hadoop-based distributed video transcoding system in cloud computing environment. International Journal of Multimedia and Ubiquitous Engineering. 2013;8(2):213–224. [Google Scholar]
- 21.Lee K, Ganti RK, Srivatsa M, Liu L. Proceedings of the 22nd ACM SIGSPATIAL International Conference on Advances in Geographic Information Systems. ACM; 2014. Efficient spatial query processing for big data; pp. 469–472. [PMC free article] [PubMed] [Google Scholar]
- 22.Li Q, Lu Y, Gong X, Zhang J. P2P, Parallel, Grid, Cloud and Internet Computing (3PGCIC), 2014 Ninth International Conference on. IEEE; 2014. Optimizational method of hbase multi-dimensional data query based on hilbert space-filling curve; pp. 469–474. [Google Scholar]
- 23.Ma T, Xu X, Tang M, Jin Y, Shen W. MHBase: A Distributed Real-Time Query Scheme for Meteorological Data Based on HBase. Future Internet. 2016;8(1):6. [Google Scholar]
- 24.Ma Y, Rao J, Hu W, Meng X, Han X, Zhang Y, Chai Y, Liu C. Proceedings of the 21st ACM international conference on Information and knowledge management. ACM; 2012. An efficient index for massive iot data in cloud environment; pp. 2129–2133. [Google Scholar]
- 25.Markonis D, Schaer R, Eggel I, Müller H, Depeursinge A. Using MapReduce for Large-scale Medical Image Analysis. arXiv preprint arXiv: 1510.06937. 2015 [Google Scholar]
- 26.McDonald C. An in-depth look at the hbase architecture. 2015 [Google Scholar]
- 27.Mohamad B, d’Orazio L, Gruenwald L. Proceedings of the 1st International Workshop on Cloud Intelligence. ACM; 2012. Towards a Hybrid Row-column Database for a Cloud-based Medical Data Management System; p. 2. [Google Scholar]
- 28.Nishimura S, Das S, Agrawal D, Abbadi AEl. 2011 IEEE 12th International Conference on Mobile Data Management. Vol. 1. IEEE; 2011. Md-hbase: A scalable multi-dimensional data infrastructure for location aware services; pp. 7–16. [Google Scholar]
- 29.Schmidt R, Rella M. International Conference on Asian Digital Libraries. Springer; 2011. An approach for processing large and nonuniform media objects on mapreduce-based clusters; pp. 172–181. [Google Scholar]
- 30.Soares TS, Dantas MA, De Macedo DD, Bauer MA. Enabling Technologies: Infrastructure for Collaborative Enterprises (WETICE), 2013 IEEE 22nd International Workshop on. IEEE; 2013. A Data Management in a Private Cloud Storage Environment Utilizing High Performance Distributed File Systems; pp. 158–163. [Google Scholar]
- 31.Sun J, Jin Q. 2010 3rd international conference on advanced computer theory and engineering (ICACTE) Vol. 1. IEEE; 2010. Scalable rdf store based on hbase and mapreduce; pp. V1–633. [Google Scholar]
- 32.Tripathy B, Mittal D. Hadoop based Uncertain Possibilistic Kernelized C-means Algorithms for Image Segmentation and a Comparative Analysis. Applied Soft Computing. 2016 [Google Scholar]
- 33.Vavilapalli VK, Murthy AC, Douglas C, Agarwal S, Konar M, Evans R, Graves T, Lowe J, Shah H, Seth S, et al. Proceedings of the 4th annual Symposium on Cloud Computing. ACM; 2013. Apache hadoop yarn: Yet another resource negotiator; p. 5. [Google Scholar]
- 34.Wang B, Li F, Hei X, Ma W, Yu L. 2015 11th International Conference on Computational Intelligence and Security (CIS) IEEE; 2015. Research on storage and retrieval method of mass data for high-speed train; pp. 474–477. [Google Scholar]
- 35.Wang S, Pandis I, Wu C, He S, Johnson D, Emam I, Guitton F, Guo Y. High dimensional biological data retrieval optimization with nosql technology. BMC genomics. 2014;15(8):1. doi: 10.1186/1471-2164-15-S8-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang CT, Shih WC, Chen LT, Kuo CT, Jiang FC, Leu FY. Accessing Medical Image File with Co-allocation HDFS in Cloud. Future Generation Computer Systems. 2015;43:61–73. [Google Scholar]


