Cildir et al. discuss the recent findings in transcriptional regulation of mast cell development and activation and provide insights into the plasticity and clinical targeting of mast cell functions.
Abstract
Mast cells are unique tissue-resident immune cells that express an array of receptors that can be activated by several extracellular cues, including antigen–immunoglobulin E (IgE) complexes, bacteria, viruses, cytokines, hormones, peptides, and drugs. Mast cells constitute a small population in tissues, but their extraordinary ability to respond rapidly by releasing granule-stored and newly made mediators underpins their importance in health and disease. In this review, we document the biology of mast cells and introduce new concepts and opinions regarding their role in human diseases beyond IgE-mediated allergic responses and antiparasitic functions. We bring to light recent discoveries and developments in mast cell research, including regulation of mast cell functions, differentiation, survival, and novel mouse models. Finally, we highlight the current and future opportunities for therapeutic intervention of mast cell functions in inflammatory diseases.
Introduction
Mast cells reside in tissues as terminally differentiated cells with a capacity to proliferate and migrate upon specific external signals. Hematopoietic stem cell (HSC)–derived mast cell progenitors circulate in blood and populate all vascularized tissues, especially at sites exposed to the external environment, including skin, conjunctiva, upper airways, lungs, and intestines (Voehringer, 2013). Mast cell differentiation, phenotype, function, and survival in these tissues are largely determined by their microenvironment, activating factors, and cytokine milieu (Galli et al., 2011).
A characteristic feature of mast cells is the presence of granules in their cytoplasm that contain a variety of biologically active molecules, including proteases (e.g., tryptase, chymase); lysosomal enzymes (e.g., cathepsins); bioactive amines (e.g., histamine, serotonin); select cytokines (e.g., TNF, IL-15); and enzymes, including granzyme B, β-hexosaminidase, and β-glucuronidase (Wernersson and Pejler, 2014). Furthermore, heparin, a proteoglycan, is a vital component of mast cell granules that is essential for the packaging of histamine and mast cell proteases (Rönnberg et al., 2012).
Notably, several cell-surface receptors are involved in mast cell activation, including high-affinity IgE receptor FcεR1 and specific G protein–coupled receptors (GPCRs) such as endothelin receptor type A (ETA), complement component 5a receptor (C5AR), adenosine A3 receptor (ADORA3), and MAS-related GPR family member X2 (MRGPRX2). Mast cells also generate cytokines, chemokines, growth factors, and lipid mediators of inflammation (e.g., prostaglandins and leukotrienes; Abraham and St. John, 2010). Therefore, equipped with a great capacity to respond to a variety of signals, mast cells have a unique and versatile role in the immune system.
Mast cells are implicated not only in allergic diseases such as asthma, hay fever, and atopic dermatitis but also in nonallergic conditions including but not limited to stroke, myocardial infarction, and cystitis. Importantly, therapeutic interventions to modulate mast cell functions can occur at several levels by blocking the action of released mediators or by preventing the transcription or release of mediators. Other targets include modulating mast cell migration, differentiation, survival, and intercellular interactions. Hence, a thorough understanding of mast cell biology and heterogeneity along with their role in human health and disease is paramount.
This review will primarily focus on the molecular mechanisms of mast cell development, activation, and functional diversity. In particular, we will highlight the recent findings in regard to the transcriptional program and signal transduction mechanisms that underlie the heterogeneity of mast cell functions. Furthermore, we will introduce the available mouse models, exploring their inherent advantages and disadvantages in investigating the biological questions in mast cell research. Finally, we will discuss the common human diseases in which mast cells are implicated and highlight new therapeutic approaches currently envisaged to target mast cell functions in the clinical setting.
Heterogeneity and evolutionary adaptations of mast cells
In humans, mast cell subtypes are defined according to whether they are positive for chymase and tryptase (MCTC) or tryptase alone (MCT). MCTC cells are comparable with mouse connective tissue mast cells, and MCT cells are comparable with mouse mucosal mast cells. The MCTC subtype is predominantly found in skin, the gastrointestinal tract, and conjunctiva, whereas the MCT subtype is the predominant mast cell type in the lungs, nose, and sinuses. However, this traditional classification is simplistic, and mast cells show significant plasticity (Galli et al., 2011). The microenvironment plays decisive roles in shaping mast cell development, phenotype, and function (Xing et al., 2011). Under basal conditions, even within the same tissue, mast cell populations are phenotypically different and form further specific subpopulations. In human lungs, for example, connective tissue mast cells in bronchi express higher levels of FcεR1 than alveolar and small-airway mast cells, suggesting location- and possibly function-dependent phenotypic alterations (Andersson et al., 2009). This location-dependent phenotypic diversity has important implications in clinical manifestations of allergic diseases. For instance, the MRGPRX2 receptor is expressed at high levels in MCTC cells in the skin but not in MCT cells in the lungs (Fujisawa et al., 2014). Therefore, activation of MRGPRX2 with diverse ligands, such as neuropeptide substance P, is associated with skin-specific effects such as itchiness.
Importantly, tissue-specific foreign microorganisms also contribute to the development and activation of mast cells, adding another layer of complexity to mast cell diversity. In mouse skin, for instance, skin microbiome can regulate the development of mast cells by inducing keratinocyte-derived stem cell factor (SCF) production, an essential growth factor for mast cell development (Wang et al., 2017). Also, in mouse skin, Staphylococcus aureus–derived δ toxin can induce mast cell activation and cause allergic skin disease (Nakamura et al., 2013), suggesting the considerable impact of the microbiome on mast cell functions.
Adipose tissue is another location where mast cells reside in the body. It is well known that mast cell progenitors are released from the bone marrow and contribute to peripheral mast cell populations. However, it is now evident that adipose tissue is also a rich source of several hematopoietic cell types, including mast cells and mast cell progenitors (Poglio et al., 2010). Notably, the numbers of adipose tissue mast cells increase upon high-fat diet (HFD)–induced obesity in mice (Cildir et al., 2013). Although mast cells were recently found to be dispensable in regulating metabolic parameters such as fat mass and insulin sensitivity in mice (Gutierrez et al., 2015; Chmelař et al., 2016), the precise functions of adipose tissue–resident mast cells have not yet been determined.
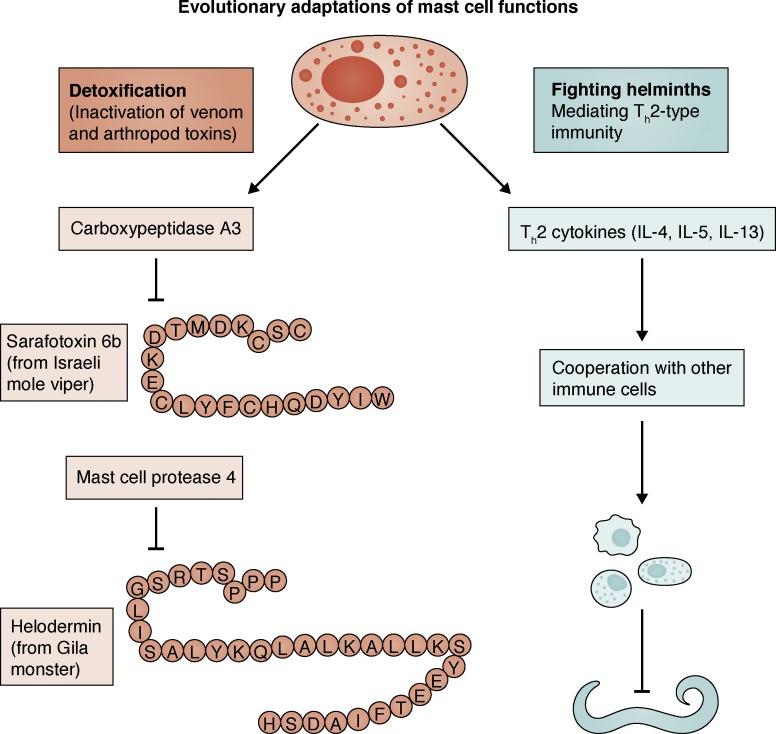
Mast cells perform diverse functions in health and disease. Their role in mounting an effective antiparasitic defense is well established (Hepworth et al., 2012; Mukai et al., 2017). For instance, in a recent study, intestinal mast cells in mice were shown to be crucial for the expulsion of intestinal helminth Heligmosomoides polygyrus (Shimokawa et al., 2017). In response to the invasion by helminth larvae, ATP is released from apoptotic intestinal epithelial cells. This activates mast cells to produce IL-33, which is important for the activation and IL-13 production of type 2 innate lymphoid cells (ILC2s), a process critical for the expulsion of helminths (Shimokawa et al., 2017). Furthermore, mast cells enhance innate and adaptive host resistance to the toxicity and mortality induced by arthropod and reptile venoms (Galli et al., 2016). In particular, mast cell–derived carboxypeptidase A3 (CPA3) and mast cell protease 4 (MCPT4) were found to inactivate toxins Sarafotoxin 6B and Helodermin, respectively (Fig. 1). Recently, mast cells were also shown to be beneficial for preserving cardiac function after myocardial infarction in mice (Ngkelo et al., 2016). In response to a myocardial infarction, mast cell progenitors from white adipose tissue were recruited to the heart and differentiated into mature mast cells. Mast cell–derived tryptase mediated muscle contractility and preserved heart function after myocardial infarction. In line with this, mast cell depletion was associated with decreased cardiac function after myocardial infarction (Ngkelo et al., 2016). Another beneficial role of mast cells has been identified in cystitis, where mouse mast cells help to eradicate uropathogenic Escherichia coli (UPEC)–infected bladder epithelial cells (BECs). After infection, BECs release IL-1β and recruit mast cells, and in turn mast cell–derived chymase is endocytosed by BECs, which induce their cytolytic death (Choi et al., 2016). It is also well known that mast cells are integral to the pathophysiology of anaphylaxis. For example, mucosal mast cells in mouse intestine contribute to food-induced intestinal anaphylaxis by being a major source of IL-9 (Chen et al., 2015). Accordingly, mice ablated for IL-9–producing mucosal mast cells had decreased food allergies. Similarly, mouse meningeal mast cells can worsen stroke pathology by releasing IL-6 and chemokine (C-C motif) ligand 7 (CCL7) and promoting the infiltration of granulocytes, brain swelling, and infarct size (Arac et al., 2014).
Figure 1.
Evolutionary adaptations of mast cell functions. Mast cell–derived mediators are critically involved in the detoxification of arthropod and reptile venoms. For example, whereas CPA3 is involved in the inhibition of Sarafotoxin 6B, MCPT4 is critical for the inhibition of Helodermin. Mast cells also contribute to the Th2-type immune response, which is critical for the prevention of helminth infections.
Although there are many examples of the heterogeneity of mast cells, in many cases the mechanistic details of this diversity remain underexamined. To gain a better understanding of the molecular mechanisms of mast cell heterogeneity and functional plasticity, it is essential to uncover the transcriptional program for mast cell development and activation.
Mast cell transcriptional program
To understand the transcriptional heterogeneity of tissue-resident mast cells, basal transcriptomes of mouse mast cells from different tissues (peritoneum, skin, esophagus, trachea, and tongue) were analyzed as part of the Immunological Genome Project (ImmGen; Dwyer et al., 2016). These results indicated that mast cells are very distantly related to the other hematopoietic cell types (Dwyer et al., 2016). Interestingly, basophils, which are widely considered to have similar functions to mast cells, exhibit a vastly different transcriptome. ImmGen results also pointed out important and surprising transcriptional differences among different tissue-resident mast cells. Notably, peritoneal and skin mast cells have the greatest transcriptional diversity. For example, the cell-surface glycoprotein CD34, an adhesion molecule previously considered to be a mast cell marker, was expressed in all mast cell types except skin mast cells. However, CD59a, a complement system regulator, had the highest expression in skin mast cells but had no expression in peritoneal mast cells in mice.
In primary human mast cells, however, transcriptomic data are more limited. As part of the Functional Annotation of the Mammalian Genome (FANTOM) project, the transcriptomes of human skin–derived mast cells have been analyzed using cap analysis of gene expression (CAGE) sequencing (Motakis et al., 2014). Similar to mouse mast cells, human skin–derived mast cells were also found to be distantly related to other immune cell types in humans. FANTOM analysis also discovered novel regulators of human mast cell functions, including bone morphogenetic protein receptor type 1A (BMPR1A). Importantly, stimulation of human mast cells with bone morphogenetic protein receptor (BMPR) ligand BMP4 prolonged mast cell survival and increased the histamine release upon FcεR1 cross-linking (Motakis et al., 2014), suggesting a cross talk between BMPRs and FcεR1 signaling in the human skin microenvironment. Overall, these results in mouse and human mast cells strongly reinforce the well-known concept of heterogeneity of mast cells in different tissues and species.
The core elements of the transcriptional program essential for mast cell development and maintenance in tissues encompass several transcription factors (TFs). These include microphthalmia-associated transcription factor (MITF), GATA binding protein 2 (GATA-2), and STAT-5, which are essential for orchestrating mast cell responses at steady state (Qi et al., 2013; Li et al., 2015). Several effector mast cell proteases and cell-surface receptors responsible for mast cell activation are transcribed by the coordinated actions of these TFs (Tshori and Nechushtan, 2012). MITF is a well-known TF in the melanocytic lineage (Levy et al., 2006), but it is also recognized as a key TF for mast cell development (Morii et al., 2004). MITF controls the expression of a diverse set of genes, including Mcpt4 and SCF receptor (Kit), which are important for mast cell functions (Shahlaee et al., 2007). Mast cells and basophils show various phenotypic similarities despite their different anatomical locations; basophils are found in circulating blood, whereas mast cells are tissue resident. Using pre-basophil and mast cell progenitors (preBMPs), CCAAT/enhancer-binding protein α (C/EBPα) has been found to be critical for basophil fate specification, whereas MITF is crucial for mast cell specification in mice (Qi et al., 2013). Notably, MITF and C/EBPα have been shown to antagonize each other’s transcriptional programs, and the expression of both C/EBPα and MITF is STAT-5 dependent (Qi et al., 2013). STAT-5 was previously shown to be important for mast cell survival in mice (Shelburne et al., 2003). In addition, STAT-5 in mast cells is required for skin inflammation, and mast cell–specific deletion of STAT-5 ameliorated atopic dermatitis in mice (Ando et al., 2014). GATA-2 is another critical factor for mast cell identity and function. GATA-2 was found to be vital for differentiation of preBMPs into basophils and mast cells in mice (Li et al., 2015). Notably, a large number of genes important for mast cell functions, including Fcer1a, Kit, and histidine decarboxylase (Hdc) are dysregulated in the absence of GATA-2. Similar to MITF, GATA-2 levels were also proposed to be regulated by STAT-5 in mast cells (Li et al., 2015), and forced expression of GATA-2 was sufficient to drive basophil and mast cell differentiation in mice. In another study in which the GATA-2 DNA-binding domain (GATA2ΔCF) was deleted in mice, BM-derived mast cell (BMMC) numbers were greatly reduced (Ohmori et al., 2015), confirming the important role of GATA-2 DNA binding and trans-activation for mast cell identity. Recently, using single-cell transcriptomic analysis of pre-granulocyte–macrophage progenitors (preGMs), GATA-1, another member of the GATA family of TFs, was also shown to be critical for marking progenitor cells generating mast cells, eosinophils, megakaryocytes, and erythroids in mice (Drissen et al., 2016). However, using tamoxifen-inducible Gata1 knockout mice (Gata1−/y), another study proposed that GATA-1, unlike GATA-2, is not essential for mast cell development in mice (Ohneda et al., 2014). Using novel mast cell–specific genetic strategies might allow us to understand the precise contribution of GATA-1 to mast cell homeostasis in future studies. Activating transcription factor 3 (ATF-3) is another TF that has been shown to influence mast cell development in mice (Gilchrist et al., 2010). ATF-3–deficient mast cells have significant developmental defects in mouse peritoneum and skin. Also, BMMCs from ATF-3–deficient mice have impaired degranulation but elevated IL-4 and IL-6 levels upon activation. However, the precise role of ATF3 in mast cell survival is not clear.
In addition to transcriptional activators, transcriptional repressors may also play an important role in mast cells. For example, hairy and enhancer of split 1 (HES1) is a repressive TF highly expressed in mast cells. HES1 is a Notch-responsive factor, and HES1 overexpression in mast cell precursors results in down-regulation of C/EBPα, a process important for mouse mast cell lineage commitment (Sakata-Yanagimoto et al., 2008). However, the precise repressive function of HES1 in mast cells is unclear and requires further investigation. In macrophages, HES1 has been described as a negative regulator of inflammation by limiting the levels of chemokine (C-X-C motif) ligand 1 (CXCL1; Shang et al., 2016), and it remains to be determined whether such regulatory mechanisms also exist in mast cells. Finally, for cell type specification, differential DNA-binding sites of TFs are crucial to drive the cell type–specific transcriptional program. It has been reported that the same TFs with similar abundance show differential binding patterns in mouse BMMCs and hematopoietic precursor cells (Calero-Nieto et al., 2014). There is also cooperativity among the TFs to drive an efficient gene expression program. The Mcpt4 locus, for example, is bound by many TFs such as GATA-2, MITF, and c-FOS cooperatively mediating its transcription (Calero-Nieto et al., 2014). Therefore, TF cooperativity seems to be a critical factor for lineage commitment and basal expression of key genes in mast cells.
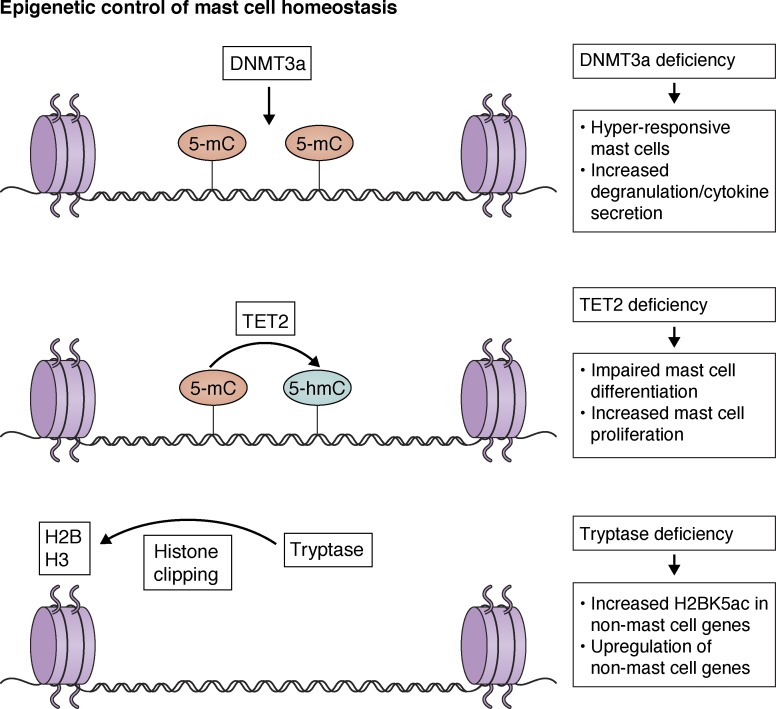
Although transcriptional analysis of mast cell development and activation is emerging, the epigenetic regulation of these processes is not well understood. In particular, how mast cell chromatin regulates mast cell functions is unclear. Recently, to address some of these questions, the functions of certain chromatin regulatory enzymes have been studied in mast cells (Fig. 2). For instance, tet methylcytosine dioxygenase 2 (TET2) is a methyl cytosine dioxygenase enzyme that converts 5-methylcytosine (5-mC) to 5-hydroxymethylcytosine (5-hmC) and thereby mediates demethylation of DNA (Rasmussen and Helin, 2016). Recently, TET2 was shown to be important for mediating mouse mast cell proliferation and in vitro differentiation (Montagner et al., 2016). Interestingly, both enzymatic activity–dependent and independent functions of TET2 have been uncovered in mast cells. Although BMMCs from TET2-deficient mice have differential 5-hmC and transcriptional profiles depending on their deoxygenase activity, increased proliferation observed in TET2-deficient mast cells was independent of its enzymatic activity, suggesting other as-yet-unknown functions of TET2 in mast cells. Another epigenetic regulator, DNA methyltransferase 3a (DNMT3a), has also been suggested to control mast cell functions (Leoni et al., 2017). As opposed to TET2, DNMT3a is a DNA methyltransferase adding methyl groups to CpG elements in DNA (Denis et al., 2011). Notably, BMMCs from DNMT3a-deficient mice are hyperresponsive to FcεR1 cross-linking, with increased degranulation and cytokine production (Leoni et al., 2017). Similarly, when reconstituted in mast cell–deficient mouse models, DNMT3a-deficient mast cells showed significantly elevated passive cutaneous anaphylaxis (PCA) responses. It is proposed that these effects, at least in part, might be due to a decrease in the levels of IQ motif-containing GTPase-activating protein 2 (IQGAP2), which regulates cytoskeleton dynamics in mast cells (Leoni et al., 2017). Another interesting epigenetic mechanism operating in mast cells is mediated by mast cell protease tryptase. Although mast cell proteases are primarily located to cytoplasmic granules, tryptase is also observed in the nucleus (Melo et al., 2014). Importantly, tryptase has been shown to catalyze the clipping of histone H2 and H3 in chromatin. In line with this, BMMCs from tryptase-deficient mice accumulate lysine 5–acetylated H2B (H2BK5ac), which also leads to induction of non–mast cell lineage genes (Melo et al., 2017). Further studies seeking to define the key mechanisms in the mast cell transcriptional program will open up opportunities in an area of potentially highly specific and effective therapeutic interventions.
Figure 2.
Epigenetic control of mast cell homeostasis. DNA methyltransferase DNMT3a is involved in de novo DNA methylation in CpG regions, which is important for gene expression regulation and homeostasis in mast cells. DNMT3a-deficient mouse mast cells are hyper-responsive to FcεR1 cross-linking and release increased mediators and cytokines. TET2, however, converts 5-mC to 5-hmC in DNA. TET2 functions are also important for mast cell proliferation and mediator release. TET2-deficient mouse mast cells are impaired in differentiation, but they have elevated proliferative capacity in culture. Importantly, TET2 mediates both enzymatic activity–dependent and independent functions in mast cells. Mast cell protease tryptase is another epigenetic regulator in mast cells. Nuclear tryptase is involved in the clipping of histone H2 and H3. Notably, tryptase deficiency results in increased H2BK5ac levels in non–mast cell genes.
Transcriptional regulation in activated mast cells
Several TFs are known to orchestrate the transcriptional response in activated mast cells for de novo synthesis of cytokines and chemokines. Similar to their roles in other cell types, NFAT, activator protein 1 (AP-1), and NF-κB have well-studied roles in driving transcription of inflammatory genes in activated mast cells (Marquardt and Walker, 2000; Lee et al., 2004; Klein et al., 2006). In addition, NF-κB signaling components have been implicated in mast cell degranulation. As an indispensable part of the NF-κB signaling pathway, IκB kinase 2 (IKK2) is an upstream kinase phosphorylating IκBα and priming its degradation to activate NF-κB subunits for DNA binding and trans-activation (Shin et al., 2014; Tong and Tergaonkar, 2014; Cildir et al., 2016). Importantly, IKK1 and IKK2 also have other cellular substrates independent of their roles in NF-κB signaling (Oeckinghaus et al., 2011). In the context of mast cell functions, it has been suggested that IKK2 is critical for phosphorylation of synaptosomal-associated protein 23 (SNAP23), an important regulator of mast cell degranulation (Lorentz et al., 2012). Because IKK2 deficiency results in embryonic lethality in mice, IKK2-deficient mouse fetal liver–derived mast cells have been generated and shown to have impaired degranulation upon FcεR1 cross-linking (Suzuki and Verma, 2008). However, in a subsequent study, connective tissue mast cell–specific deletion of IKK2 (Mcpt5-Cre, IKK2F/F mice) resulted in normal degranulation but impaired cytokine production from peritoneal-derived mast cells (Peschke et al., 2014). In the same study, Mcpt5-Cre–mediated deletion of NF-κB essential modulator (NEMO; another essential subunit of the IKK complex mediating NF-κB activation) also resulted in impaired cytokine production from activated mouse peritoneal mast cells. Finally, using BMS-345541, a pharmacological inhibitor of IKK2 kinase activity, a recent study proposed that IKK2 is important for SNAP23 and syntaxin-4 (STX4) complex formation and cytokine production upon FcεR1 cross-linking (Gaudenzio et al., 2016). Overall, it is possible that the role of IKK2 in mast cell degranulation is context dependent, but IKK2 is undeniably essential for the synthesis of a subset of cytokines in activated mast cells by controlling NF-κB activation.
Early growth response 1 (EGR1) and EGR2 are early-response TFs that are members of the zinc finger TF family and are induced by a diverse array of extracellular stimuli in different cell types (Seiler et al., 2012). In mouse BMMCs, de novo synthesized EGR1 has been shown to regulate TNF and IL-13 production upon FcεR1 cross-linking (Li et al., 2006, 2007). A similar TF, EGR2, however, has been shown to regulate chemokine (C-C motif) ligand 1 (CCL1) production in mouse fetal liver–derived mast cells, which is important for the recruitment of T cells into inflamed tissues (Wu et al., 2013). Although the binding of EGR1 and EGR2 has been suggested to occur in the promoter regions of certain proinflammatory genes, it is, however, important to identify the genome-wide binding patterns of EGR1 and EGR2 to obtain a complete picture of their effect on activated mast cells. Zinc finger E-box–binding homeobox 2 (ZEB2) is another zinc finger TF that is implicated in the activation of BMMCs based on siRNA-mediated knockdown experiments. Although it has been shown that ZEB2 is highly expressed in mast cells and knocking down ZEB2 levels caused impaired activation of mast cells upon FcεR1 cross-linking (Barbu et al., 2012), its genome-wide DNA-binding pattern in mast cells and genetic deficiency in mice require further investigation.
In conclusion, our understanding of the transcriptional dynamics of mast cell development and activation is limited, and most studies are still based on mouse mast cells. Because TFs are critical for the expression of cell-surface receptors and extracellular mediators involved in mast cell activation, strategies specifically targeting mast cell TFs could be developed and tested in future clinical studies.
Modes of mast cell activation
Although it is well known that mast cells can be activated by different ligand–receptor complexes, it has only recently become evident that there are notable differences between different modes of mast cell activation. The recent identification of the MRGPRX2 receptor and its mouse orthologue Mrgprb2, which are activated by several ligands such as neuropeptide substance P, and several pseudoallergic drugs has opened up a new research avenue and introduced a new therapeutic target for the control of pseudoallergic reactions (McNeil et al., 2015). It has been shown that Mrgprb2 is specifically expressed on connective tissue mast cells but not on mucosal mast cells in mice, and the activation of Mrgprb2 and MRGPRX2 by diverse ligands leads to mast cell degranulation (McNeil et al., 2015).
A disease relevance for MRGPRX2 has also started to emerge; the levels of MRGPRX2 expression on skin mast cells have been observed to be higher in chronic urticaria patients compared with healthy individuals (Fujisawa et al., 2014), suggesting dynamic regulation of its surface expression on mast cells. Recently, an elegant study using large chemical libraries and structural modeling discovered novel agonists of MRGPRX2 (Lansu et al., 2017). These agonists include many opioid compounds and peptides such as (–) and (+) morphine; hydrocodone; sinomenine; dynorphin A; dynorphin B; and a synthetic compound, ZINC-3573, which has very high specificity toward MRGPRX2 but not to other GPCRs. Importantly, these compounds caused mast cell degranulation in human mast cells in an MRGPRX2-dependent manner (Lansu et al., 2017). Overall, these findings have important implications for humans; morphine and similar analgesics are known to induce histamine release and itching in humans (Baldo and Pham, 2012). However, whether the effects of opioid ligands on mast cells are also physiologically relevant should be tested in vivo using nonhuman primate models. MRGPRX2 and its mouse orthologue Mrgprb2 have only a 52% sequence homology, and MRGPRX2 is conserved only in primates. In line with this, opioid compounds did not show a significant effect on the Mrgprb2 receptor (Lansu et al., 2017).
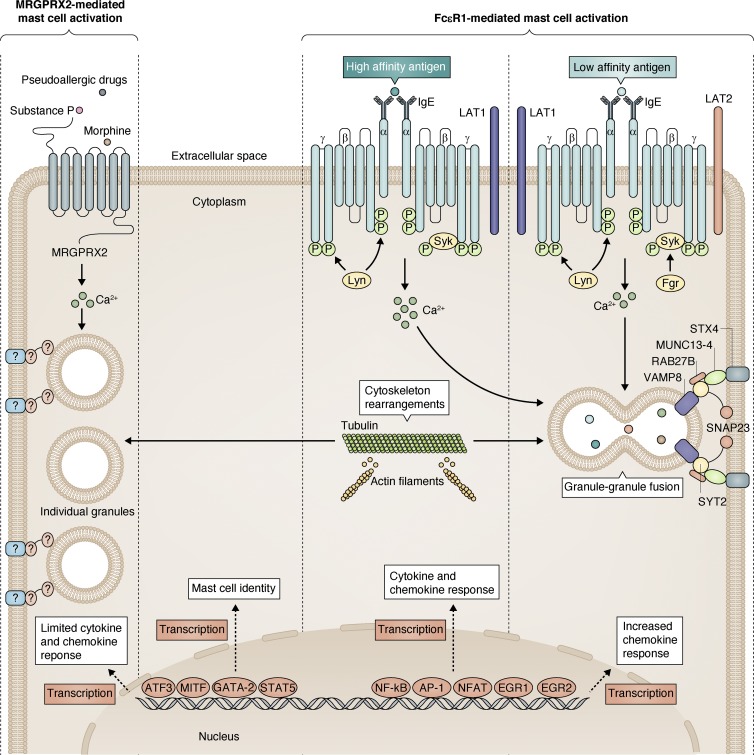
It has now become evident that there are important differences between MRGPRX2- and FcεR1-mediated mast cell activation and degranulation (Fig. 3). Classically, IgE-dependent mast cell activation occurs via FcεR1 comprising four polypeptide chains: an α, a β, and two γ (Galli and Tsai, 2012). The β and γ chains have an immunoreceptor tyrosine-based activation motif (ITAM) that is essential for signal transduction mediated by Lck/Yes novel tyrosine kinase (Lyn) and spleen tyrosine kinase (Syk) phosphorylations, whereas the extracellular domains of the α subunit bind to IgE (Blank et al., 2014). Immediately after antigen- and IgE-induced cross-linking of cell-surface FcεR1, activation of the kinases Lyn and Syk leads to the rapid phosphorylation of FcεR1 ITAMs and the subsequent activation of multiple downstream events, including increased cytosolic calcium levels and cytoskeleton rearrangements such as microtubule polymerization and actin depolymerization (Dráber et al., 2012; Fig. 3). Importantly, FcεR1-mediated mast cell activation involves a robust inflammatory response with newly made cytokines and chemokines, mediated by several TFs such as NFAT, AP-1, NF-κB, EGR1, and EGR2, and this response has been shown to be limited in MRGPRX2-mediated activation (Gaudenzio et al., 2016). Although FcεR1 cross-linking results in granule–granule fusions in cytoplasm before granule release, MRGPRX2 activation results in more uniform and rapid release of individual granules (Gaudenzio et al., 2016). Furthermore, FcεR1-mediated granule release is known to be orchestrated by several soluble N-ethylmaleimide–sensitive factor-activating protein receptor (SNARE) complexes and adaptor proteins, including but not limited to SNAP23, STX4, vesicle-associated membrane protein 8 (VAMP8), protein Unc-13 homologue D (MUNC 13-4), and Ras-related protein Rab-27B (RAB27B; Lorentz et al., 2012; Fig. 3), but the identity of these complexes is less well known in MRGPRX2-mediated degranulation. Finally, depending on the affinity of the antigen to IgE, FcεR1 cross-linking causes differential cellular and physiological effects in mast cells. Importantly, the strength of, and qualitative changes in, the molecular signals downstream of FcεR1 can be discriminated by high-affinity or low-affinity antigen stimulus despite equivalent FcεR1 phosphorylation (Suzuki et al., 2014). In particular, low-affinity antigens activate feline Gardner–Rasheed sarcoma viral oncogene homologue (Fgr) kinase and shift the balance from linker for activation of T cells (LAT1) adaptor molecules toward LAT2 adaptor molecules, which then initiates a more chemokine-based inflammatory response compared with the high-affinity antigens (Suzuki et al., 2014; Fig. 3). Consistent with the higher intracellular calcium increase after FcεR1 cross-linking, high-affinity antigen binding also results in higher degranulation (Suzuki et al., 2014).
Figure 3.
Different modes of mast cell activation. Mast cell identity is maintained by the coordinated actions of several TFs such as MITF, GATA-2, STAT-5, and ATF-3. Mast cells can be activated by different GPCRs such as MRGPRX2 and high-affinity IgE receptor FcεR1. However, there are notable differences between MRGPRX2- and FcεR1-mediated mast cell activation. Although ligand binding to MRGPRX2 leads to intracellular calcium increase and degranulation, there is very limited cytokine response. However, high-affinity antigen-mediated FcεR1 cross-linking involves higher intracellular calcium levels, degranulation, and inflammatory cytokine and chemokine response involving various TFs such as NF-κB, NFAT, AP-1, EGR1, and EGR2. Despite similar levels of FcεR1 receptor phosphorylation, low-affinity antigen-mediated FcεR1 cross-linking involves a smaller increase in intracellular calcium levels and degranulation, and the inflammatory response is more chemokine based compared with high-affinity antigen-mediated activation. This is due to the involvement of cytoplasmic Fgr kinase and the membrane-bound LAT2 adaptor. Finally, although cytoskeleton rearrangements such as microtubule polymerization and actin depolymerization are a common feature of all forms of degranulation, the components of SNARE machinery are less well known in MRGPRX2-mediated degranulation.
IgE binding to the FcεR1 complex is a major mechanism for activation of mast cells. However, it is uncertain how tissue-resident mast cells acquire IgE molecules from the bloodstream. By studying reporter IgE molecules, it has been demonstrated that tissue-resident perivascular mast cells acquire IgE by extending their protrusions across the blood vessel walls (Cheng et al., 2013). This indicates that IgE uptake is a dynamic process and that perivascular positioning of mast cells is important to sample IgE molecules in blood (Cheng et al., 2013).
It is important to note that coactivation of various cell-surface receptors can enhance or diminish FcεR1 signaling in mast cells. For example, T cell immunoglobulin and mucin-domain containing-3 (TIM3) is a cell-surface immune checkpoint regulator that is also expressed on mast cells (Phong et al., 2015). Importantly, cross-linking of the TIM3 receptor with specific antibodies simultaneously with FcεR1 cross-linking resulted in increased mast cell degranulation and cytokine secretion (Phong et al., 2015). Similarly, BMMCs from TIM3-deficient mice have been reported to have reduced activation-induced cytokine levels, suggesting positive regulatory functions of TIM3 in FcεR1 signaling (Phong et al., 2015). Sphingosine-1-phosphate receptor 2 (S1PR2) has also been proposed to regulate FcεR1 signaling, and S1PR2 antagonists significantly reduce mast cell degranulation and cytokine secretion upon FcεR1 cross-linking (Oskeritzian et al., 2010). In addition, TNF receptor superfamily member 14 (TNFRSF14) activation by its ligand cytokine TNFSF14 (LIGHT) has been reported to enhance FcεR1 signaling and result in enhanced degranulation in both human mast cells and mouse BMMCs (Sibilano et al., 2016). By binding to IL-6R, IL-6 increases the reactivity of human mast cells to FcεR1 cross-linking by regulating the levels of STAT-3 and suppressor of cytokine signaling 3 (SOCS3; Desai et al., 2016). Similarly, by binding to IL33R/ST2, IL-33 pretreatment increases the IgE-mediated degranulation and cytokine secretion in human mast cells (Joulia et al., 2017). In contrast, co-ligation of the allergy-inhibitory receptor (Allergin-1), an Ig-like receptor, and FcεR1 impairs degranulation in mouse BMMCs (Hitomi et al., 2010). A novel mechanism of inhibition of mast cell responses was recently proposed. Referred to as trans-inhibition, it is mediated by phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase 1 (SHIP1; Malbec et al., 2016). It was previously known that inhibitory receptor FcγRIIB blocks mast cell activation when it is co-engaged with FcεR1 (cis-inhibition). However, in trans-inhibition, it is proposed that receptors recruiting sufficient amounts of SHIP1 phosphatase, including but not limited to FcγRIIB, can inhibit FcεR1-mediated mast cell activation and Kit-dependent mast cell proliferation (Malbec et al., 2016). Overall, it is conceivable that in future studies other receptors that regulate FcεR1- and MRGPRX2-dependent mast cell activation could be identified and targeted to alleviate allergic inflammation.
New mouse models of mast cells
In vivo animal models have greatly enhanced our understanding of mast cell functions. Many novel mouse models with different genetic strategies have recently been developed and used to test the previously proposed functions of mast cells and gain novel mechanistic insights. The functions of mast cells and mast cell–derived mediators have historically been analyzed using mouse models in which cell surface receptor KIT has been disrupted. Initially, these included mouse models WBB6F1-KitW/W-v and C57BL/6-KitW-sh/W-sh. These models use either point mutations, causing the Kit protein to be unable to be expressed on the cell surface (Kitw) and limiting its kinase activity (KitW-v), or an inversion mutation (KitW-sh) in upstream regulatory regions of the Kit locus in mice (Reber et al., 2012). Notably, both of these KIT-dependent mouse models use adoptive transfer of BMMCs into tissues, and there are concerns as to how closely adoptively transferred mast cells resemble native populations in tissues. Moreover, the route of BMMC injection, the number of injected cells, and the analysis time after the injection could also contribute to the variations in mast cell responses in these models. In addition, it is now evident that these models affect non–mast cell populations as well, thereby introducing inherent confounding variables in mast cell research. For example, WBB6F1-KitW/W-v mice are anemic and sterile. They also have several abnormalities reported in other cell types, including decreased numbers of basophils and neutrophils, defects in interstitial cells of Cajal, melanocytes, and γδ T cells in the gut (Reber et al., 2012). However, whereas C57BL/6-KitW-sh/W-sh mice are fertile and not anemic, they show defects in melanocytes, reduced levels of IgE, and elevated numbers of neutrophils and basophils (Reber et al., 2012).
To circumvent some of the aforementioned problems in WBB6F1-KitW/W-v and C57BL/6-KitW-sh/W-sh mouse models, new KIT-independent mouse models have been generated to study mast cell functions. These new mouse models could lead to constitutive or inducible mast cell deficiency or allow mast cell–specific inactivation of genes of interest. Importantly, some findings on the roles of mast cells using KIT-dependent mouse models have been refuted using KIT-independent mouse models (Feyerabend et al., 2011; Gutierrez et al., 2015).
KIT-independent mouse models with constitutive mast cell deficiency include Mcpt5-Cre; R-DTA mice (in which the diphtheria toxin [DT] α chain is expressed in Mcpt5-expressing mast cells), Cpa3Cre/+ mice (also known as “Cre-Master,” heterozygous mice in which Cre is expressed under the Cpa3 promoter), and Cpa3-Cre; Mcl-1F/F mice (also known as “Hello Kitty” mice in which the antiapoptotic gene Mcl-1 is deleted in Cpa3-expressing mast cells). Furthermore, mouse models with inducible depletion of mast cells include Mcpt5-Cre; iDTR mice (in which Mcpt5-Cre mice crossed with iDTRF/F mice, thereby allowing Cre-dependent expression of DTR in Mcpt5-expressing mast cells and depletion of these cells upon DT injection), Mas-TRECK mice (in which human DTR is expressed under the control of IL-4 intronic enhancer elements), and RMB mice (in which DTR and bright red td-Tomato [tdT] fluorescent proteins are expressed in 3′ UTR of the membrane spanning 4-domains A2 [MS4A2] gene encoding for the FcεR β chain; Reber et al., 2012; Dahdah et al., 2014). In addition, mast cell–specific gene deletion has been achieved with Mcpt5-Cre, Cpa3-Cre, and FcεR1β-Cre mouse models (Furumoto et al., 2011; Lilla et al., 2011; Peschke et al., 2014). However, some of these new mouse models also have specific disadvantages or undesired Cre activities in other cell populations. For instance, Mcpt5-Cre mice enable gene inactivation only in connective tissue mast cells but not in mucosal mast cells, and Cre activity in this model has also been reported in a small fraction of NK cells in blood (Dudeck et al., 2011). Similarly, although the Cpa3-Cre model enables gene inactivation in both mucosal mast cells and connective tissue mast cells, Cpa3 is also expressed by basophils, some populations of T cell progenitors, thymic T cells, and hematopoietic progenitor cells. In line with this, “Hello Kitty” mice also have defects in non–mast cell populations and show macrocytic anemia (Lilla et al., 2011). Apart from these models, it has been shown that genetic deficiency of several genes could also result in impaired mast cell development in mice (Table 1).
Table 1. Selected examples of genes important for mast cell development.
| Gene | Mouse model | Phenotype | Reference |
|---|---|---|---|
| GATA2 | GATA2ΔCF | Loss of mast cell phenotype in BMMCs | Ohmori et al., 2015 |
| STAT5 | STAT5A/B +/− | Deficiency of mast cells in all analyzed tissues | Shelburne et al., 2003 |
| MITF | MITF −/− (tg/tg mice) | Deficiency of mast cells in peritoneum, mesentery, stomach, spleen | Morii et al., 2004 |
| NDST2 | NDST2 −/− | Decreased numbers of connective tissue mast cells and reduced mast cell proteases and histamine content | Forsberg et al., 1999 |
| HDC | HDC −/− mice | Decreased numbers of peritoneal and mesenterial mast cells and decreased histamine content | Ohtsu et al., 2001 |
| CPA3 | CPA3 −/− mice | Immature peritoneal mast cells with reduced granular staining | Feyerabend et al., 2005 |
| PI3K | p85α −/− and p110γ −/− DKO mice | Severe defects in mast cell development | Takayama et al., 2013 |
| PI3K | p85α −/− mice | Mast cell deficiency in gastrointestinal and peritoneal mast cells | Fukao et al., 2002 |
| AHR | AHR −/− mice | Reduction in skin, peritoneal cavity, and stomach mucosa–resident mast cells | Zhou et al., 2013 |
| SRGN | SRGN −/− mice | Deficiency in granulated mast cells in peritoneum and ear | Åbrink et al., 2004 |
| PLA2G3 | PLA2G3 −/− mice | Irregularly shaped and immature tissue mast cells | Taketomi et al., 2013 |
| Mac-1 (CD11b/CD18, CR3) | Mac-1 −/− mice | Decreased number of mast cells in peritoneum and dorsal epidermis | Rosenkranz et al., 1998 |
| P38α | Mx-Cre+ P38αF/F (P38α−/−) | Decreased numbers of peritoneal and lung mast cells | Hu et al., 2012 |
Finally, although mouse models have greatly contributed to our understanding of mouse mast cells, the contribution of human mast cells to in vivo responses has been difficult to analyze. Therefore, a humanized mouse model (NSG-SGM3 BLT mice) has recently been developed (Bryce et al., 2016). This model provides a rich source of human mast cells that could be activated by chimeric IgE molecules and could be used to develop in vivo disease models in which therapeutics may be tested.
Mast cell–mediated human diseases
The involvement of mast cells in the progression of many human diseases is becoming increasingly clear, and many therapeutics are being developed to control abnormal mast cell activation and proliferation in several inflammatory diseases, including allergic asthma, rhinitis, conjunctivitis, dermatitis, food allergies, chronic rhinosinusitis, nasal polyps, aspirin-exacerbated respiratory disease (AERD), and chronic urticaria (Galli and Tsai, 2012). In these conditions, mast cells, together with other immune cells such as eosinophils, macrophages, and T cells, are involved in disease progression. These diseases affect both adults and children, and the prevalence of allergic diseases is estimated to be 19.6% of the population. In 2007, the financial cost of allergies was estimated to be $6 billion USD (Beggs et al., 2015).
Primary mast cell dysfunction results in mastocytosis and mast cell activation disorders (MCADs; Ustun et al., 2016). Whereas mastocytosis involves the aberrant accumulation of mast cells in many tissues and organs, MCADs involve the chronic, uncontrolled activation of mast cells. Importantly, several causative mutations have been identified in mastocytosis patients, KIT D816V being the most common. KIT D816V mutation leads to autoactivation of KIT receptor tyrosine kinase, which then constitutively drives mast cell proliferation (Valent et al., 2017). Additional KIT mutations (such as KIT D419H and M541L) and mutations in other genes (such as c-CBL, TET2, SRSF2, ASXL1, and RUNX1) have also recently been identified in mastocytosis patients (Schwaab et al., 2013; Jawhar et al., 2016). Although it is unknown if these mutations are drivers or passengers, their coexistence with common KIT mutations may lead to increased proliferation or hyperactivation of mast cells and is a marker for poor prognosis in mastocytosis patients (Jawhar et al., 2016). Therefore, it is important to determine which specific non-KIT mutations also predispose individuals to mastocytosis. Given the facts that epigenetic regulators DNMT3a and TET2 have been shown to modulate mast cell proliferation in mice (Montagner et al., 2016; Leoni et al., 2017) and that DNMT3a and TET2 mutations are also frequent in human mastocytosis patients (Traina et al., 2012), it is also imperative to determine whether these mutations affect the expression or catalytic functions of TET2 and DNMT3a.
Apart from mast cell clonal diseases and activation disorders, some patients show abnormal levels of mast cell–derived mediators in basal conditions. Because mast cell proteases are secreted upon mast cell activation, their abnormal levels in blood serum is also a significant risk factor for developing inflammatory disorders. It has been shown that basal serum levels of tryptase are higher in some individuals without showing signs of mastocytosis (Fellinger et al., 2014). Using genome sequencing to account for the genetic cause of elevated basal tryptase levels in a cohort of patients, germline duplications and triplications in TPSAB1 gene coding for α-tryptase protein have recently been identified (Lyons et al., 2016). These patients with higher basal tryptase levels show various clinical features, including joint hypermobility, skeletal abnormalities, body pain, and headaches. Importantly, peripheral blood–derived mast cells generated from these patients exhibit more spontaneous tryptase secretion (Lyons et al., 2016).
Novel approaches in clinical targeting of mast cell functions
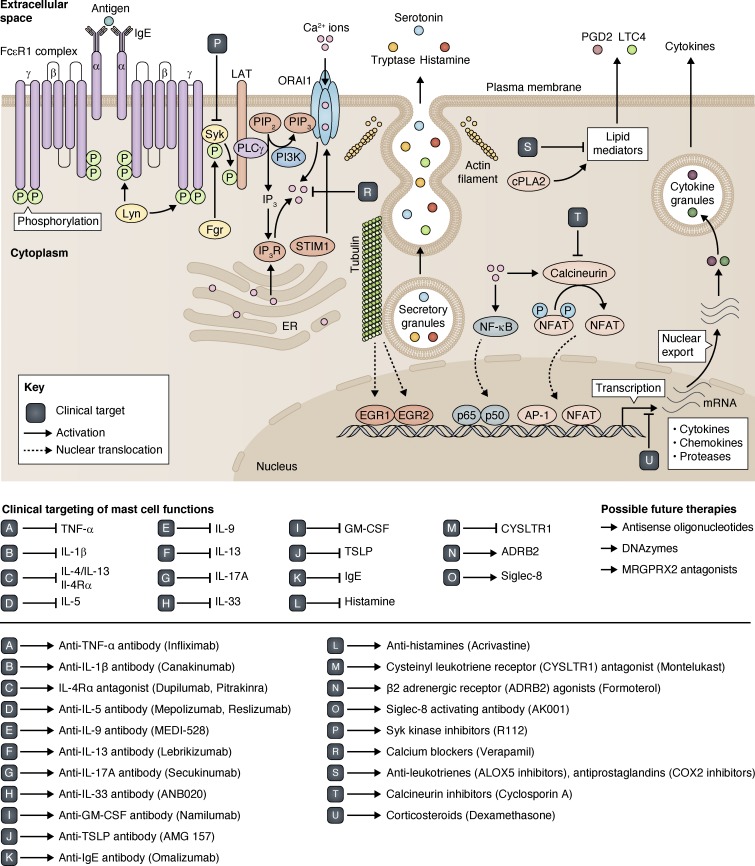
The actions of mast cell–derived mediators contribute to the pathophysiology of common allergic disorders. In clinical settings, various strategies targeting intracellular or extracellular mast cell mediators have traditionally been tested in allergic patients, and some of them are in common clinical use (Fig. 4). However, some of these treatments are only partially effective, have sedative side effects, are not tolerated by all patients, or are considered safe to use long term in children. For instance, corticosteroids (e.g., oral prednisolone, topical budesonide and fluticasone) suppress some of the IgE-mediated activities and provide partial to complete short-term symptom control. However, some patients are resistant to the effect of corticosteroids, whereas others develop corticosteroid resistance over the course of their disease (often those with severe asthma or nasal polyps; Aversa et al., 2012; Barnes, 2013). Furthermore, oral corticosteroids are not safe for all patients, especially those with peptic ulcer disease or a history of shingles, and their long-term use has numerous well-known side effects (Narum et al., 2014). Antihistamines also have variable efficacy and sedative side effects. Some newer therapies have been recently designed to improve the efficacy profile in patients (Akdis, 2012). For example, Omalizumab is a humanized monoclonal antibody that binds to free IgE in circulation, acting as a competitive inhibitor for binding to FcεR1 (Normansell et al., 2014). It is an FDA-approved drug currently in use to treat severe asthma and chronic idiopathic urticaria. Furthermore, anti–IL-5 monoclonal antibody (e.g., Mepolizumab) has also shown promising results in patients with severe asthma (Ortega et al., 2014). Similarly, newer biologics such as anti–IL-4/anti–IL-13 monoclonal antibody (Dupilumab) are showing better efficacy profiles compared with corticosteroid alone in atopic dermatitis, chronic rhinosinusitis, and nasal polyps (Thaçi et al., 2016; Wenzel et al., 2016). This highlights the notion that multiple therapeutic targets are required to control chronic inflammation.
Figure 4.
Clinical targeting of mast cell functions. There are various extracellular cytokines that are released from mast cells or that regulate the mast cell responses after binding to their corresponding receptors on mast cells. Some of these cytokines have been targeted using blocking monoclonal antibodies, which are in use to treat allergic disorders such as asthma. These cytokines include TNF, IL-1, IL-5, IL-9, IL-13, IL-17A, IL-33, GM-CSF, and thymic stromal lymphopoietin (TSLP). Furthermore, extracellular IgE is targeted with Omalizumab, and histamine is targeted with antihistamines. Similarly, many cell-surface receptors involved in mast cell activation are also therapeutic targets. These receptors include IL-4Rα (a common receptor for IL-4 and IL-13), cysteinyl leukotriene receptor 1 (CYSLTR1), and β-2 adrenergic receptor (ADRB2). Intracellular targets in mast cells include Syk kinase (mediating LAT phosphorylation and signal propagation), intracellular calcium, arachidonate 5–lipoxygenase (ALOX5; important for leukotriene synthesis), cyclooxygenase 2 (COX-2; important for prostaglandin synthesis), calcineurin (phosphatase activating NFAT TF), and the glucocorticoid receptor (inhibiting proinflammatory cytokine synthesis). Inhibitory receptor Siglec 8 is also another therapeutic target. Promising future therapies might also include the use of antisense oligonucleotides, DNAzymes, and small-molecule inhibitors of MRGPRX2 receptor activation.
Targeting upstream molecules controlling the activation of numerous proinflammatory downstream mediators is an attractive therapeutic approach. For example, GATA-3 is a critical TF controlling the expression of Th2 genes, which is a common signature in asthma. Traditionally, TFs were considered to be difficult to target in vivo, but deoxyribozyme (DNAzyme) technology has promise in this field of therapeutics. Notably, an inhaled GATA-3–specific DNAzyme, SB010, has been shown to be effective in cleaving and inactivating GATA-3 mRNAs, in turn reducing mRNA and protein levels of GATA-3. More importantly, SB010 has demonstrated considerable efficacy in clinical trials with allergic asthma patients (Krug et al., 2015). Similar approaches might also be beneficial to target upstream regulators of mast cell functions in other mast cell–related diseases, including mastocytosis and MCADs.
Use of antisense oligonucleotides is another promising approach for therapeutic targeting of mast cell functions. For example, MS4A2 loci coding for the β chain of FcεR1 (FcεR1β) is associated with allergies in humans (Kim et al., 2006) and, in a recent study, a truncated variant of FcεR1β (t-FcεR1β) was also identified in human mast cells (Cruse et al., 2010). T-FcεR1β lacks exon 3 of MS4A2, coding for the first two transmembrane domains of FcεR1β; therefore, t-FcεR1β does not traffic to the plasma membrane (Cruse et al., 2013). Antisense oligonucleotide–mediated exon skipping has been successfully applied to specifically manipulate MS4A2 splicing, target FcεR1β, and down-regulate the expression and activation of FcεR1 (Cruse et al., 2016). Notably, local applications of antisense oligonucleotides have the potential to control mast cell responses in allergic disorders. Another promising therapeutic target for allergic diseases is the MRGPRX2 receptor. Because of its specific expression pattern on connective tissue mast cells (apart from some neurons), targeting MRGPRX2 receptor activation with small-molecule inhibitors or monoclonal antibodies might alleviate common pseudoallergic reactions.
Inhibitory receptors such as sialic acid binding Ig-like lectin (Siglec) receptors represent further novel therapeutic targets in mast cells. Although there are many Siglec receptors with different expression patterns in immune cells, Siglec 8 is specifically expressed in human mast cells and eosinophils, and pretreatment of human mast cells generated from CD34+ precursors with an antibody activating Siglec 8 has been shown to inhibit mast cell degranulation (Kiwamoto et al., 2012). Importantly, clinical trials with antibodies activating Siglec 8 (AK001 and AK002) are ongoing in nasal polyposis and systemic mastocytosis patients. Similarly, Siglec 7 has also been shown to dampen FcεR1-dependent mast cell activation (Mizrahi et al., 2014), and antibodies activating Siglec 7 could also be a viable therapeutic option in the future.
Concluding remarks and future directions
Mast cells play an increasingly recognized role in health and disease. Recent developments in mast cell research have reinforced the understanding that mast cells have unique immune functions, a deeper understanding of which will have a great impact on human health. As new genomic tools and mouse models become more available, our understanding of mast cell functions and plasticity will continue to increase. However, there are still several challenges to be addressed for better control of mast cell activation and proliferation in disease settings.
First of all, mouse and human mast cells may have functional differences, and findings on mouse mast cells should be supported by studies on human mast cells wherever possible. In particular, we believe that humanized mouse models will be a valuable tool to investigate the functions of human mast cells in vivo. Second, with the recent advances in cutting-edge techniques such as CRISPR, single-cell sequencing, and CyTOF, it is now feasible to identify novel molecular targets and delineate previously unknown genetic and epigenetic mechanisms in human mast cells. For instance, single-cell sequencing shows great promise in uncovering subpopulations of various tissue-resident cells, and it could be applied to delineate specific subpopulations of mast cells in different human tissues in steady state as well as in acute and chronic inflammatory settings. Furthermore, we still have a very limited understanding of the roles of noncoding RNAs in mast cell development or activation. Many studies have shown that noncoding RNAs play an important role in controlling inflammatory responses, and it is conceivable that long noncoding RNAs might also play a previously unknown role in mast cell development or activation. Loss-of-function studies using CRISPR in human mast cells will help determine the unknown roles of RNAs in mast cell biology.
Because mast cells generate many different cytokines, chemokines, and other soluble mediators, targeting an upstream regulator rather than a single mediator will undoubtedly be a more viable approach in controlling allergic diseases. In this regard, further research on the MRGPRX2 receptor will provide important clues to control pseudoallergic reactions commonly seen in the population.
Finally, it is now widely accepted that the microbiome has immense effects on human physiology and that many diseases have connections with tissue-specific microbiomes. Because mast cells also reside in the skin and intestines, the effects of the skin and gut microbiome on mast cell functions should be further investigated.
Outstanding questions
What are the specific extracellular cues and intracellular transcription factors regulating MRGPRX2 transcription in mast cells, and how does MRGPRX2 structurally recognize diverse ligands?
MRGPRX2 is expressed on connective tissue mast cells but not on mucosal mast cells, suggesting that specific extracellular signals and associated transcription factors might be involved in inducing the transcription of MRGPRX2. MRGPRX2 could be activated by numerous ligands, including pseudoallergic drugs, neuropeptides, and opioid compounds. Detailed analysis of MRGPRX2–ligand interactions responsible for mast cell activation must be performed for therapeutic purposes.
Are there any polymorphisms in coding regions or cis-regulatory domains of the human MRGPRX2 locus?
MRGPRX2 variants might predispose individuals to hyperactivation by changing the structure of MRGPRX2. Similarly, single-nucleotide polymorphisms (SNPs) in cis-regulatory domains of the MRGPRX2 locus might regulate the expression of the receptor on cells.
Which common non-KIT mutations are drivers of mastocytosis?
Although several common mutations have been observed in mastocytosis patients, it is unknown if they are driver or passenger mutations. CRISPR/Cas9-based genome editing will allow testing of the involvement of these mutations in mast cell proliferation.
Could DNAzyme technology be applied to intracellular mast cell targets?
Given the fact that the GATA-3–specific DNAzyme has shown efficacy in asthma patients, it is also worth testing specific DNAzymes for intracellular targets in mast cells.
Acknowledgments
The authors would like to thank Stephen J. Galli, Stephanie A. Conos, and Simon Conn for their comments on the manuscript.
The authors also gratefully acknowledge funding provided by the Agency for Science, Technology and Research (A*STAR) of Singapore; the National Health and Medical Research Council (NHMRC) of Australia; and the Premier’s Research and Industry Fund grant provided by the South Australian Government Department of State Development.
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- BMMC
- BM-derived mast cell
- DT
- diphtheria toxin
- GPCR
- G protein–coupled receptor
- MCAD
- mast cell activation disorder
- MITF
- microphthalmia-associated transcription factor
- SCF
- stem cell factor
- TF
- transcription factor
References
- Abraham S.N., and St. John A.L.. 2010. Mast cell-orchestrated immunity to pathogens. Nat. Rev. Immunol. 10:440–452. 10.1038/nri2782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Åbrink M., Grujic M., and Pejler G.. 2004. Serglycin is essential for maturation of mast cell secretory granule. J. Biol. Chem. 279:40897–40905. 10.1074/jbc.M405856200 [DOI] [PubMed] [Google Scholar]
- Akdis C.A. 2012. Therapies for allergic inflammation: Refining strategies to induce tolerance. Nat. Med. 18:736–749. 10.1038/nm.2754 [DOI] [PubMed] [Google Scholar]
- Andersson C.K., Mori M., Bjermer L., Löfdahl C.G., and Erjefält J.S.. 2009. Novel site-specific mast cell subpopulations in the human lung. Thorax. 64:297–305. 10.1136/thx.2008.101683 [DOI] [PubMed] [Google Scholar]
- Ando T., Xiao W., Gao P., Namiranian S., Matsumoto K., Tomimori Y., Hong H., Yamashita H., Kimura M., Kashiwakura J., et al. . 2014. Critical role for mast cell Stat5 activity in skin inflammation. Cell Reports. 6:366–376. 10.1016/j.celrep.2013.12.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arac A., Grimbaldeston M.A., Nepomuceno A.R., Olayiwola O., Pereira M.P., Nishiyama Y., Tsykin A., Goodall G.J., Schlecht U., Vogel H., et al. . 2014. Evidence that meningeal mast cells can worsen stroke pathology in mice. Am. J. Pathol. 184:2493–2504. 10.1016/j.ajpath.2014.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aversa S., Ondolo C., Abbadessa G., Piccione F., Carriero V., Fulcheri A., Lauria A., De Francia S., and Racca S.. 2012. Steroid resistance in nasal polyposis: Role of glucocorticoid receptor and TGF-β1. Rhinology. 50:427–435. [DOI] [PubMed] [Google Scholar]
- Baldo B.A., and Pham N.H.. 2012. Histamine-releasing and allergenic properties of opioid analgesic drugs: Resolving the two. Anaesth. Intensive Care. 40:216–235. [DOI] [PubMed] [Google Scholar]
- Barbu E.A., Zhang J., Berenstein E.H., Groves J.R., Parks L.M., and Siraganian R.P.. 2012. The transcription factor Zeb2 regulates signaling in mast cells. J. Immunol. 188:6278–6286. 10.4049/jimmunol.1102660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes P.J. 2013. Corticosteroid resistance in patients with asthma and chronic obstructive pulmonary disease. J. Allergy Clin. Immunol. 131:636–645. 10.1016/j.jaci.2012.12.1564 [DOI] [PubMed] [Google Scholar]
- Beggs P.J., Katelaris C.H., Medek D., Johnston F.H., Burton P.K., Campbell B., Jaggard A.K., Vicendese D., Bowman D.M., Godwin I., et al. . 2015. Differences in grass pollen allergen exposure across Australia. Aust. N. Z. J. Public Health. 39:51–55. 10.1111/1753-6405.12325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blank U., Madera-Salcedo I.K., Danelli L., Claver J., Tiwari N., Sánchez-Miranda E., Vázquez-Victorio G., Ramírez-Valadez K.A., Macias-Silva M., and González-Espinosa C.. 2014. Vesicular trafficking and signaling for cytokine and chemokine secretion in mast cells. Front. Immunol. 5:453 10.3389/fimmu.2014.00453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryce P.J., Falahati R., Kenney L.L., Leung J., Bebbington C., Tomasevic N., Krier R.A., Hsu C.L., Shultz L.D., Greiner D.L., and Brehm M.A.. 2016. Humanized mouse model of mast cell–mediated passive cutaneous anaphylaxis and passive systemic anaphylaxis. J. Allergy Clin. Immunol. 138:769–779. 10.1016/j.jaci.2016.01.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calero-Nieto F.J., Ng F.S., Wilson N.K., Hannah R., Moignard V., Leal-Cervantes A.I., Jimenez-Madrid I., Diamanti E., Wernisch L., and Göttgens B.. 2014. Key regulators control distinct transcriptional programmes in blood progenitor and mast cells. EMBO J. 33:1212–1226. 10.1002/embj.201386825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.Y., Lee J.B., Liu B., Ohta S., Wang P.Y., Kartashov A.V., Mugge L., Abonia J.P., Barski A., Izuhara K., et al. . 2015. Induction of interleukin-9-producing mucosal mast cells promotes susceptibility to IgE-mediated experimental food allergy. Immunity. 43:788–802. 10.1016/j.immuni.2015.08.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L.E., Hartmann K., Roers A., Krummel M.F., and Locksley R.M.. 2013. Perivascular mast cells dynamically probe cutaneous blood vessels to capture immunoglobulin E. Immunity. 38:166–175. 10.1016/j.immuni.2012.09.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chmelař J., Chatzigeorgiou A., Chung K.J., Prucnal M., Voehringer D., Roers A., and Chavakis T.. 2016. No role for mast cells in obesity-related metabolic dysregulation. Front. Immunol. 7:524 10.3389/fimmu.2016.00524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H.W., Bowen S.E., Miao Y., Chan C.Y., Miao E.A., Abrink M., Moeser A.J., and Abraham S.N.. 2016. Loss of bladder epithelium induced by cytolytic mast cell granules. Immunity. 45:1258–1269. 10.1016/j.immuni.2016.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cildir G., Akıncılar S.C., and Tergaonkar V.. 2013. Chronic adipose tissue inflammation: All immune cells on the stage. Trends Mol. Med. 19:487–500. 10.1016/j.molmed.2013.05.001 [DOI] [PubMed] [Google Scholar]
- Cildir G., Low K.C., and Tergaonkar V.. 2016. Noncanonical NF-κB signaling in health and disease. Trends Mol. Med. 22:414–429. 10.1016/j.molmed.2016.03.002 [DOI] [PubMed] [Google Scholar]
- Cruse G., Kaur D., Leyland M., and Bradding P.. 2010. A novel FcεRIβ-chain truncation regulates human mast cell proliferation and survival. FASEB J. 24:4047–4057. 10.1096/fj.10-158378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruse G., Beaven M.A., Ashmole I., Bradding P., Gilfillan A.M., and Metcalfe D.D.. 2013. A truncated splice-variant of the FcεRIβ receptor subunit is critical for microtubule formation and degranulation in mast cells. Immunity. 38:906–917. 10.1016/j.immuni.2013.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruse G., Yin Y., Fukuyama T., Desai A., Arthur G.K., Bäumer W., Beaven M.A., and Metcalfe D.D.. 2016. Exon skipping of FcεRIβ eliminates expression of the high-affinity IgE receptor in mast cells with therapeutic potential for allergy. Proc. Natl. Acad. Sci. USA. 113:14115–14120. 10.1073/pnas.1608520113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahdah A., Gautier G., Attout T., Fiore F., Lebourdais E., Msallam R., Daëron M., Monteiro R.C., Benhamou M., Charles N., et al. . 2014. Mast cells aggravate sepsis by inhibiting peritoneal macrophage phagocytosis. J. Clin. Invest. 124:4577–4589. 10.1172/JCI75212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denis H., Ndlovu M.N., and Fuks F.. 2011. Regulation of mammalian DNA methyltransferases: A route to new mechanisms. EMBO Rep. 12:647–656. 10.1038/embor.2011.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai A., Jung M.Y., Olivera A., Gilfillan A.M., Prussin C., Kirshenbaum A.S., Beaven M.A., and Metcalfe D.D.. 2016. IL-6 promotes an increase in human mast cell numbers and reactivity through suppression of suppressor of cytokine signaling 3. J. Allergy Clin. Immunol. 137:1863–1871.e6. 10.1016/j.jaci.2015.09.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dráber P., Sulimenko V., and Dráberová E.. 2012. Cytoskeleton in mast cell signaling. Front. Immunol. 3:130 10.3389/fimmu.2012.00130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drissen R., Buza-Vidas N., Woll P., Thongjuea S., Gambardella A., Giustacchini A., Mancini E., Zriwil A., Lutteropp M., Grover A., et al. . 2016. Distinct myeloid progenitor-differentiation pathways identified through single-cell RNA sequencing. Nat. Immunol. 17:666–676. 10.1038/ni.3412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudeck A., Dudeck J., Scholten J., Petzold A., Surianarayanan S., Köhler A., Peschke K., Vöhringer D., Waskow C., Krieg T., et al. . 2011. Mast cells are key promoters of contact allergy that mediate the adjuvant effects of haptens. Immunity. 34:973–984. 10.1016/j.immuni.2011.03.028 [DOI] [PubMed] [Google Scholar]
- Dwyer D.F., Barrett N.A., and Austen K.F.. Immunological Genome Project Consortium . 2016. Expression profiling of constitutive mast cells reveals a unique identity within the immune system. Nat. Immunol. 17:878–887. 10.1038/ni.3445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellinger C., Hemmer W., Wöhrl S., Sesztak-Greinecker G., Jarisch R., and Wantke F.. 2014. Clinical characteristics and risk profile of patients with elevated baseline serum tryptase. Allergol. Immunopathol. (Madr.). 42:544–552. 10.1016/j.aller.2014.05.002 [DOI] [PubMed] [Google Scholar]
- Feyerabend T.B., Hausser H., Tietz A., Blum C., Hellman L., Straus A.H., Takahashi H.K., Morgan E.S., Dvorak A.M., Fehling H.J., and Rodewald H.R.. 2005. Loss of histochemical identity in mast cells lacking carboxypeptidase A. Mol. Cell. Biol. 25:6199–6210. 10.1128/MCB.25.14.6199-6210.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feyerabend T.B., Weiser A., Tietz A., Stassen M., Harris N., Kopf M., Radermacher P., Möller P., Benoist C., Mathis D., et al. . 2011. Cre-mediated cell ablation contests mast cell contribution in models of antibody- and T cell-mediated autoimmunity. Immunity. 35:832–844. 10.1016/j.immuni.2011.09.015 [DOI] [PubMed] [Google Scholar]
- Forsberg E., Pejler G., Ringvall M., Lunderius C., Tomasini-Johansson B., Kusche-Gullberg M., Eriksson I., Ledin J., Hellman L., and Kjellén L.. 1999. Abnormal mast cells in mice deficient in a heparin-synthesizing enzyme. Nature. 400:773–776. 10.1038/23488 [DOI] [PubMed] [Google Scholar]
- Fujisawa D., Kashiwakura J., Kita H., Kikukawa Y., Fujitani Y., Sasaki-Sakamoto T., Kuroda K., Nunomura S., Hayama K., Terui T., et al. . 2014. Expression of Mas-related gene X2 on mast cells is upregulated in the skin of patients with severe chronic urticaria. J. Allergy Clin. Immunol. 134:622–633.e9. 10.1016/j.jaci.2014.05.004 [DOI] [PubMed] [Google Scholar]
- Fukao T., Yamada T., Tanabe M., Terauchi Y., Ota T., Takayama T., Asano T., Takeuchi T., Kadowaki T., Hata Ji J., and Koyasu S.. 2002. Selective loss of gastrointestinal mast cells and impaired immunity in PI3K-deficient mice. Nat. Immunol. 3:295–304. 10.1038/ni768 [DOI] [PubMed] [Google Scholar]
- Furumoto Y., Charles N., Olivera A., Leung W.H., Dillahunt S., Sargent J.L., Tinsley K., Odom S., Scott E., Wilson T.M., et al. . 2011. PTEN deficiency in mast cells causes a mastocytosis-like proliferative disease that heightens allergic responses and vascular permeability. Blood. 118:5466–5475. 10.1182/blood-2010-09-309955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli S.J., and Tsai M.. 2012. IgE and mast cells in allergic disease. Nat. Med. 18:693–704. 10.1038/nm.2755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli S.J., Borregaard N., and Wynn T.A.. 2011. Phenotypic and functional plasticity of cells of innate immunity: Macrophages, mast cells and neutrophils. Nat. Immunol. 12:1035–1044. 10.1038/ni.2109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli S.J., Starkl P., Marichal T., and Tsai M.. 2016. Mast cells and IgE in defense against venoms: Possible “good side” of allergy? Allergol. Int. 65:3–15. 10.1016/j.alit.2015.09.002 [DOI] [PubMed] [Google Scholar]
- Gaudenzio N., Sibilano R., Marichal T., Starkl P., Reber L.L., Cenac N., McNeil B.D., Dong X., Hernandez J.D., Sagi-Eisenberg R., et al. . 2016. Different activation signals induce distinct mast cell degranulation strategies. J. Clin. Invest. 126:3981–3998. 10.1172/JCI85538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist M., Henderson W.R. Jr., Morotti A., Johnson C.D., Nachman A., Schmitz F., Smith K.D., and Aderem A.. 2010. A key role for ATF3 in regulating mast cell survival and mediator release. Blood. 115:4734–4741. 10.1182/blood-2009-03-213512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez D.A., Muralidhar S., Feyerabend T.B., Herzig S., and Rodewald H.R.. 2015. Hematopoietic kit deficiency, rather than lack of mast cells, protects mice from obesity and insulin resistance. Cell Metab. 21:678–691. 10.1016/j.cmet.2015.04.013 [DOI] [PubMed] [Google Scholar]
- Hepworth M.R., Daniłowicz-Luebert E., Rausch S., Metz M., Klotz C., Maurer M., and Hartmann S.. 2012. Mast cells orchestrate type 2 immunity to helminths through regulation of tissue-derived cytokines. Proc. Natl. Acad. Sci. USA. 109:6644–6649. 10.1073/pnas.1112268109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitomi K., Tahara-Hanaoka S., Someya S., Fujiki A., Tada H., Sugiyama T., Shibayama S., Shibuya K., and Shibuya A.. 2010. An immunoglobulin-like receptor, Allergin-1, inhibits immunoglobulin E–mediated immediate hypersensitivity reactions. Nat. Immunol. 11:601–607. 10.1038/ni.1886 [DOI] [PubMed] [Google Scholar]
- Hu P., Carlesso N., Xu M., Liu Y., Nebreda A.R., Takemoto C., and Kapur R.. 2012. Genetic evidence for critical roles of P38α protein in regulating mast cell differentiation and chemotaxis through distinct mechanisms. J. Biol. Chem. 287:20258–20269. 10.1074/jbc.M112.358119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jawhar M., Schwaab J., Schnittger S., Meggendorfer M., Pfirrmann M., Sotlar K., Horny H.P., Metzgeroth G., Kluger S., Naumann N., et al. . 2016. Additional mutations in SRSF2, ASXL1 and/or RUNX1 identify a high risk group of patients with KIT D816V+ advanced systemic mastocytosis. Leukemia. 30:136–143. 10.1038/leu.2015.284 [DOI] [PubMed] [Google Scholar]
- Joulia R., L’Faqihi F.E., Valitutti S., and Espinosa E.. 2017. IL-33 fine tunes mast cell degranulation and chemokine production at the single-cell level. J. Allergy Clin. Immunol. 140:497–509.e10. 10.1016/j.jaci.2016.09.049 [DOI] [PubMed] [Google Scholar]
- Kim S.H., Bae J.S., Holloway J.W., Lee J.T., Suh C.H., Nahm D.H., and Park H.S.. 2006. A polymorphism of MS4A2 (−109T>C) encoding the β-chain of the high-affinity immunoglobulin E receptor (FcεR1β) is associated with a susceptibility to aspirin-intolerant asthma. Clin. Exp. Allergy. 36:877–883. 10.1111/j.1365-2222.2006.02443.x [DOI] [PubMed] [Google Scholar]
- Kiwamoto T., Kawasaki N., Paulson J.C., and Bochner B.S.. 2012. Siglec-8 as a drugable target to treat eosinophil and mast cell-associated conditions. Pharmacol. Ther. 135:327–336. 10.1016/j.pharmthera.2012.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein M., Klein-Hessling S., Palmetshofer A., Serfling E., Tertilt C., Bopp T., Heib V., Becker M., Taube C., Schild H., et al. . 2006. Specific and redundant roles for NFAT transcription factors in the expression of mast cell-derived cytokines. J. Immunol. 177:6667–6674. 10.4049/jimmunol.177.10.6667 [DOI] [PubMed] [Google Scholar]
- Krug N., Hohlfeld J.M., Kirsten A.M., Kornmann O., Beeh K.M., Kappeler D., Korn S., Ignatenko S., Timmer W., Rogon C., et al. . 2015. Allergen-induced asthmatic responses modified by a GATA3-specific DNAzyme. N. Engl. J. Med. 372:1987–1995. 10.1056/NEJMoa1411776 [DOI] [PubMed] [Google Scholar]
- Lansu K., Karpiak J., Liu J., Huang X.P., McCorvy J.D., Kroeze W.K., Che T., Nagase H., Carroll F.I., Jin J., et al. . 2017. In silico design of novel probes for the atypical opioid receptor MRGPRX2. Nat. Chem. Biol. 13:529–536. 10.1038/nchembio.2334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.N., Tuckerman J., Nechushtan H., Schutz G., Razin E., and Angel P.. 2004. c-Fos as a regulator of degranulation and cytokine production in FcεRI-activated mast cells. J. Immunol. 173:2571–2577. 10.4049/jimmunol.173.4.2571 [DOI] [PubMed] [Google Scholar]
- Leoni C., Montagner S., Rinaldi A., Bertoni F., Polletti S., Balestrieri C., and Monticelli S.. 2017. Dnmt3a restrains mast cell inflammatory responses. Proc. Natl. Acad. Sci. USA. 114:E1490–E1499. 10.1073/pnas.1616420114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy C., Khaled M., and Fisher D.E.. 2006. MITF: Master regulator of melanocyte development and melanoma oncogene. Trends Mol. Med. 12:406–414. 10.1016/j.molmed.2006.07.008 [DOI] [PubMed] [Google Scholar]
- Li B., Power M.R., and Lin T.J.. 2006. De novo synthesis of early growth response factor-1 is required for the full responsiveness of mast cells to produce TNF and IL-13 by IgE and antigen stimulation. Blood. 107:2814–2820. 10.1182/blood-2005-09-3610 [DOI] [PubMed] [Google Scholar]
- Li B., Berman J., Tang J.T., and Lin T.J.. 2007. The early growth response factor-1 is involved in stem cell factor (SCF)-induced interleukin 13 production by mast cells, but is dispensable for SCF-dependent mast cell growth. J. Biol. Chem. 282:22573–22581. 10.1074/jbc.M610859200 [DOI] [PubMed] [Google Scholar]
- Li Y., Qi X., Liu B., and Huang H.. 2015. The STAT5-GATA2 pathway is critical in basophil and mast cell differentiation and maintenance. J. Immunol. 194:4328–4338. 10.4049/jimmunol.1500018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilla J.N., Chen C.C., Mukai K., BenBarak M.J., Franco C.B., Kalesnikoff J., Yu M., Tsai M., Piliponsky A.M., and Galli S.J.. 2011. Reduced mast cell and basophil numbers and function in Cpa3-Cre; Mcl-1fl/fl mice. Blood. 118:6930–6938. 10.1182/blood-2011-03-343962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorentz A., Baumann A., Vitte J., and Blank U.. 2012. The SNARE machinery in mast cell secretion. Front. Immunol. 3:143 10.3389/fimmu.2012.00143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons J.J., Yu X., Hughes J.D., Le Q.T., Jamil A., Bai Y., Ho N., Zhao M., Liu Y., O’Connell M.P., et al. . 2016. Elevated basal serum tryptase identifies a multisystem disorder associated with increased TPSAB1 copy number. Nat. Genet. 48:1564–1569. 10.1038/ng.3696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malbec O., Cassard L., Albanesi M., Jönsson F., Mancardi D., Chicanne G., Payrastre B., Dubreuil P., Vivier E., and Daëron M.. 2016. Trans-inhibition of activation and proliferation signals by Fc receptors in mast cells and basophils. Sci. Signal. 9:ra126 10.1126/scisignal.aag1401 [DOI] [PubMed] [Google Scholar]
- Marquardt D.L., and Walker L.L.. 2000. Dependence of mast cell IgE-mediated cytokine production on nuclear factor-κB activity. J. Allergy Clin. Immunol. 105:500–505. 10.1067/mai.2000.104942 [DOI] [PubMed] [Google Scholar]
- McNeil B.D., Pundir P., Meeker S., Han L., Undem B.J., Kulka M., and Dong X.. 2015. Identification of a mast-cell-specific receptor crucial for pseudo-allergic drug reactions. Nature. 519:237–241. 10.1038/nature14022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo F.R., Vita F., Berent-Maoz B., Levi-Schaffer F., Zabucchi G., and Pejler G.. 2014. Proteolytic histone modification by mast cell tryptase, a serglycin proteoglycan-dependent secretory granule protease. J. Biol. Chem. 289:7682–7690. 10.1074/jbc.M113.546895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo F.R., Wallerman O., Paivandy A., Calounova G., Gustafson A.M., Sabari B.R., Zabucchi G., Allis C.D., and Pejler G.. 2017. Tryptase-catalyzed core histone truncation: A novel epigenetic regulatory mechanism in mast cells. J. Allergy Clin. Immunol. 140:474–485. 10.1016/j.jaci.2016.11.044 [DOI] [PubMed] [Google Scholar]
- Mizrahi S., Gibbs B.F., Karra L., Ben-Zimra M., and Levi-Schaffer F.. 2014. Siglec-7 is an inhibitory receptor on human mast cells and basophils. J. Allergy Clin. Immunol. 134:230–233.e3. 10.1016/j.jaci.2014.03.031 [DOI] [PubMed] [Google Scholar]
- Montagner S., Leoni C., Emming S., Della Chiara G., Balestrieri C., Barozzi I., Piccolo V., Togher S., Ko M., Rao A., et al. . 2016. TET2 regulates mast cell differentiation and proliferation through catalytic and non-catalytic activities. Cell Reports. 15:1566–1579. 10.1016/j.celrep.2016.04.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morii E., Oboki K., Ishihara K., Jippo T., Hirano T., and Kitamura Y.. 2004. Roles of MITF for development of mast cells in mice: Effects on both precursors and tissue environments. Blood. 104:1656–1661. 10.1182/blood-2004-01-0247 [DOI] [PubMed] [Google Scholar]
- Motakis E., Guhl S., Ishizu Y., Itoh M., Kawaji H., de Hoon M., Lassmann T., Carninci P., Hayashizaki Y., Zuberbier T., et al. . 2014. Redefinition of the human mast cell transcriptome by deep-CAGE sequencing. Blood. 123:e58–e67. 10.1182/blood-2013-02-483792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai K., Karasuyama H., Kabashima K., Kubo M., and Galli S.J.. 2017. Differences in the importance of mast cells, basophils, IgE, and IgG versus that of CD4+ T cells and ILC2 cells in primary and secondary immunity to Strongyloides venezuelensis. Infect. Immun. 85:e00053-17 10.1128/IAI.00053-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y., Oscherwitz J., Cease K.B., Chan S.M., Muñoz-Planillo R., Hasegawa M., Villaruz A.E., Cheung G.Y., McGavin M.J., Travers J.B., et al. . 2013. Staphylococcus δ-toxin induces allergic skin disease by activating mast cells. Nature. 503:397–401. 10.1038/nature12655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narum S., Westergren T., and Klemp M.. 2014. Corticosteroids and risk of gastrointestinal bleeding: A systematic review and meta-analysis. BMJ Open. 4:e004587 10.1136/bmjopen-2013-004587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngkelo A., Richart A., Kirk J.A., Bonnin P., Vilar J., Lemitre M., Marck P., Branchereau M., Le Gall S., Renault N., et al. . 2016. Mast cells regulate myofilament calcium sensitization and heart function after myocardial infarction. J. Exp. Med. 213:1353–1374. 10.1084/jem.20160081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normansell R., Walker S., Milan S.J., Walters E.H., and Nair P.. 2014. Omalizumab for asthma in adults and children. Cochrane Database Syst. Rev. 13:CD003559 10.1002/14651858.CD003559.pub4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeckinghaus A., Hayden M.S., and Ghosh S.. 2011. Crosstalk in NF-κB signaling pathways. Nat. Immunol. 12:695–708. 10.1038/ni.2065 [DOI] [PubMed] [Google Scholar]
- Ohmori S., Moriguchi T., Noguchi Y., Ikeda M., Kobayashi K., Tomaru N., Ishijima Y., Ohneda O., Yamamoto M., and Ohneda K.. 2015. GATA2 is critical for the maintenance of cellular identity in differentiated mast cells derived from mouse bone marrow. Blood. 125:3306–3315. 10.1182/blood-2014-11-612465 [DOI] [PubMed] [Google Scholar]
- Ohneda K., Moriguchi T., Ohmori S., Ishijima Y., Satoh H., Philipsen S., and Yamamoto M.. 2014. Transcription factor GATA1 is dispensable for mast cell differentiation in adult mice. Mol. Cell. Biol. 34:1812–1826. 10.1128/MCB.01524-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsu H., Tanaka S., Terui T., Hori Y., Makabe-Kobayashi Y., Pejler G., Tchougounova E., Hellman L., Gertsenstein M., Hirasawa N., et al. . 2001. Mice lacking histidine decarboxylase exhibit abnormal mast cells. FEBS Lett. 502:53–56. 10.1016/S0014-5793(01)02663-1 [DOI] [PubMed] [Google Scholar]
- Ortega H.G., Liu M.C., Pavord I.D., Brusselle G.G., FitzGerald J.M., Chetta A., Humbert M., Katz L.E., Keene O.N., Yancey S.W., et al. . 2014. Mepolizumab treatment in patients with severe eosinophilic asthma. N. Engl. J. Med. 371:1198–1207. 10.1056/NEJMoa1403290 [DOI] [PubMed] [Google Scholar]
- Oskeritzian C.A., Price M.M., Hait N.C., Kapitonov D., Falanga Y.T., Morales J.K., Ryan J.J., Milstien S., and Spiegel S.. 2010. Essential roles of sphingosine-1-phosphate receptor 2 in human mast cell activation, anaphylaxis, and pulmonary edema. J. Exp. Med. 207:465–474. 10.1084/jem.20091513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschke K., Weitzmann A., Heger K., Behrendt R., Schubert N., Scholten J., Voehringer D., Hartmann K., Dudeck A., Schmidt-Supprian M., and Roers A.. 2014. IκB kinase 2 is essential for IgE-induced mast cell de novo cytokine production but not for degranulation. Cell Reports. 8:1300–1307. 10.1016/j.celrep.2014.07.046 [DOI] [PubMed] [Google Scholar]
- Phong B.L., Avery L., Sumpter T.L., Gorman J.V., Watkins S.C., Colgan J.D., and Kane L.P.. 2015. Tim-3 enhances FcεRI-proximal signaling to modulate mast cell activation. J. Exp. Med. 212:2289–2304. 10.1084/jem.20150388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poglio S., De Toni-Costes F., Arnaud E., Laharrague P., Espinosa E., Casteilla L., and Cousin B.. 2010. Adipose tissue as a dedicated reservoir of functional mast cell progenitors. Stem Cells. 28:2065–2072. 10.1002/stem.523 [DOI] [PubMed] [Google Scholar]
- Qi X., Hong J., Chaves L., Zhuang Y., Chen Y., Wang D., Chabon J., Graham B., Ohmori K., Li Y., and Huang H.. 2013. Antagonistic regulation by the transcription factors C/EBPα and MITF specifies basophil and mast cell fates. Immunity. 39:97–110. 10.1016/j.immuni.2013.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen K.D., and Helin K.. 2016. Role of TET enzymes in DNA methylation, development, and cancer. Genes Dev. 30:733–750. 10.1101/gad.276568.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reber L.L., Marichal T., and Galli S.J.. 2012. New models for analyzing mast cell functions in vivo. Trends Immunol. 33:613–625. 10.1016/j.it.2012.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rönnberg E., Melo F.R., and Pejler G.. 2012. Mast cell proteoglycans. J. Histochem. Cytochem. 60:950–962. 10.1369/0022155412458927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenkranz A.R., Coxon A., Maurer M., Gurish M.F., Austen K.F., Friend D.S., Galli S.J., and Mayadas T.N.. 1998. Impaired mast cell development and innate immunity in Mac-1 (CD11b/CD18, CR3)-deficient mice. J. Immunol. 161:6463–6467. [PubMed] [Google Scholar]
- Sakata-Yanagimoto M., Nakagami-Yamaguchi E., Saito T., Kumano K., Yasutomo K., Ogawa S., Kurokawa M., and Chiba S.. 2008. Coordinated regulation of transcription factors through Notch2 is an important mediator of mast cell fate. Proc. Natl. Acad. Sci. USA. 105:7839–7844. 10.1073/pnas.0801074105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwaab J., Schnittger S., Sotlar K., Walz C., Fabarius A., Pfirrmann M., Kohlmann A., Grossmann V., Meggendorfer M., Horny H.P., et al. . 2013. Comprehensive mutational profiling in advanced systemic mastocytosis. Blood. 122:2460–2466. 10.1182/blood-2013-04-496448 [DOI] [PubMed] [Google Scholar]
- Seiler M.P., Mathew R., Liszewski M.K., Spooner C.J., Barr K., Meng F., Singh H., and Bendelac A.. 2012. Elevated and sustained expression of the transcription factors Egr1 and Egr2 controls NKT lineage differentiation in response to TCR signaling. Nat. Immunol. 13:264–271. 10.1038/ni.2230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahlaee A.H., Brandal S., Lee Y.N., Jie C., and Takemoto C.M.. 2007. Distinct and shared transcriptomes are regulated by microphthalmia-associated transcription factor isoforms in mast cells. J. Immunol. 178:378–388. 10.4049/jimmunol.178.1.378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y., Coppo M., He T., Ning F., Yu L., Kang L., Zhang B., Ju C., Qiao Y., Zhao B., et al. . 2016. The transcriptional repressor Hes1 attenuates inflammation by regulating transcription elongation. Nat. Immunol. 17:930–937. 10.1038/ni.3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelburne C.P., McCoy M.E., Piekorz R., Sexl V., Roh K.H., Jacobs-Helber S.M., Gillespie S.R., Bailey D.P., Mirmonsef P., Mann M.N., et al. . 2003. Stat5 expression is critical for mast cell development and survival. Blood. 102:1290–1297. 10.1182/blood-2002-11-3490 [DOI] [PubMed] [Google Scholar]
- Shimokawa C., Kanaya T., Hachisuka M., Ishiwata K., Hisaeda H., Kurashima Y., Kiyono H., Yoshimoto T., Kaisho T., and Ohno H.. 2017. Mast cells are crucial for induction of group 2 innate lymphoid cells and clearance of helminth infections. Immunity. 46:863–874.e4. 10.1016/j.immuni.2017.04.017 [DOI] [PubMed] [Google Scholar]
- Shin E.M., Hay H.S., Lee M.H., Goh J.N., Tan T.Z., Sen Y.P., Lim S.W., Yousef E.M., Ong H.T., Thike A.A., et al. . 2014. DEAD-box helicase DP103 defines metastatic potential of human breast cancers. J. Clin. Invest. 124:3807–3824. 10.1172/JCI73451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibilano R., Gaudenzio N., DeGorter M.K., Reber L.L., Hernandez J.D., Starkl P.M., Zurek O.W., Tsai M., Zahner S., Montgomery S.B., et al. . 2016. A TNFRSF14-FcεRI-mast cell pathway contributes to development of multiple features of asthma pathology in mice. Nat. Commun. 7:13696 10.1038/ncomms13696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K., and Verma I.M.. 2008. Phosphorylation of SNAP-23 by IκB kinase 2 regulates mast cell degranulation. Cell. 134:485–495. 10.1016/j.cell.2008.05.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki R., Leach S., Liu W., Ralston E., Scheffel J., Zhang W., Lowell C.A., and Rivera J.. 2014. Molecular editing of cellular responses by the high-affinity receptor for IgE. Science. 343:1021–1025. 10.1126/science.1246976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama G., Ohtani M., Minowa A., Matsuda S., and Koyasu S.. 2013. Class I PI3K-mediated Akt and ERK signals play a critical role in FcεRI-induced degranulation in mast cells. Int. Immunol. 25:215–220. 10.1093/intimm/dxs105 [DOI] [PubMed] [Google Scholar]
- Taketomi Y., Ueno N., Kojima T., Sato H., Murase R., Yamamoto K., Tanaka S., Sakanaka M., Nakamura M., Nishito Y., et al. . 2013. Mast cell maturation is driven via a group III phospholipase A2-prostaglandin D2–DP1 receptor paracrine axis. Nat. Immunol. 14:554–563. 10.1038/ni.2586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaçi D., Simpson E.L., Beck L.A., Bieber T., Blauvelt A., Papp K., Soong W., Worm M., Szepietowski J.C., Sofen H., et al. . 2016. Efficacy and safety of dupilumab in adults with moderate-to-severe atopic dermatitis inadequately controlled by topical treatments: A randomised, placebo-controlled, dose-ranging phase 2b trial. Lancet. 387:40–52. 10.1016/S0140-6736(15)00388-8 [DOI] [PubMed] [Google Scholar]
- Tong L., and Tergaonkar V.. 2014. Rho protein GTPases and their interactions with NFκB: crossroads of inflammation and matrix biology. Biosci. Rep. 34:e00115 10.1042/BSR20140021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traina F., Visconte V., Jankowska A.M., Makishima H., O’Keefe C.L., Elson P., Han Y., Hsieh F.H., Sekeres M.A., Mali R.S., et al. . 2012. Single nucleotide polymorphism array lesions, TET2, DNMT3A, ASXL1 and CBL mutations are present in systemic mastocytosis. PLoS One. 7:e43090 10.1371/journal.pone.0043090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tshori S., and Nechushtan H.. 2012. Mast cell transcription factors—Regulators of cell fate and phenotype. Biochim. Biophys. Acta. 1822:42–48. 10.1016/j.bbadis.2010.12.024 [DOI] [PubMed] [Google Scholar]
- Ustun C., Arock M., Kluin-Nelemans H.C., Reiter A., Sperr W.R., George T., Horny H.P., Hartmann K., Sotlar K., Damaj G., et al. . 2016. Advanced systemic mastocytosis: From molecular and genetic progress to clinical practice. Haematologica. 101:1133–1143. 10.3324/haematol.2016.146563 [DOI] [PubMed] [Google Scholar]
- Valent P., Akin C., Hartmann K., Nilsson G., Reiter A., Hermine O., Sotlar K., Sperr W.R., Escribano L., George T.I., et al. . 2017. Advances in the classification and treatment of mastocytosis: Current status and outlook toward the future. Cancer Res. 77:1261–1270. 10.1158/0008-5472.CAN-16-2234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voehringer D. 2013. Protective and pathological roles of mast cells and basophils. Nat. Rev. Immunol. 13:362–375. 10.1038/nri3427 [DOI] [PubMed] [Google Scholar]
- Wang Z., Mascarenhas N., Eckmannm L., Miyamoto Y., Sun X., Kawakami T., and Di Nardo A.. 2017. Skin microbiome promotes mast cell maturation by triggering stem cell factor production in keratinocytes. J. Allergy Clin. Immunol. 139:1205–1216.e6. 10.1016/j.jaci.2016.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel S., Castro M., Corren J., Maspero J., Wang L., Zhang B., Pirozzi G., Sutherland E.R., Evans R.R., Joish V.N., et al. . 2016. Dupilumab efficacy and safety in adults with uncontrolled persistent asthma despite use of medium-to-high-dose inhaled corticosteroids plus a long-acting β2 agonist: A randomised double-blind placebo-controlled pivotal phase 2b dose-ranging trial. Lancet. 388:31–44. 10.1016/S0140-6736(16)30307-5 [DOI] [PubMed] [Google Scholar]
- Wernersson S., and Pejler G.. 2014. Mast cell secretory granules: Armed for battle. Nat. Rev. Immunol. 14:478–494. 10.1038/nri3690 [DOI] [PubMed] [Google Scholar]
- Wu Z., MacNeil A.J., Junkins R., Li B., Berman J.N., and Lin T.J.. 2013. Mast cell FcεRI-induced early growth response 2 regulates CC chemokine ligand 1–dependent CD4+ T cell migration. J. Immunol. 190:4500–4507. 10.4049/jimmunol.1203158 [DOI] [PubMed] [Google Scholar]
- Xing W., Austen K.F., Gurish M.F., and Jones T.G.. 2011. Protease phenotype of constitutive connective tissue and of induced mucosal mast cells in mice is regulated by the tissue. Proc. Natl. Acad. Sci. USA. 108:14210–14215. 10.1073/pnas.1111048108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Tung H.Y., Tsai Y.M., Hsu S.C., Chang H.W., Kawasaki H., Tseng H.C., Plunkett B., Gao P., Hung C.H., et al. . 2013. Aryl hydrocarbon receptor controls murine mast cell homeostasis. Blood. 121:3195–3204. 10.1182/blood-2012-08-453597 [DOI] [PMC free article] [PubMed] [Google Scholar]