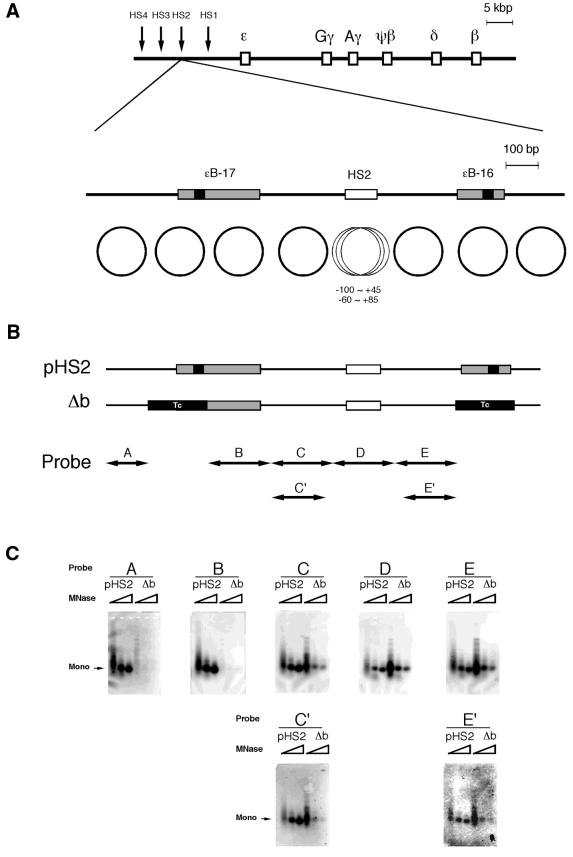
Figure 1.
Nucleosomal phases in the presence or absence of curved DNA. (A) Maps of the human β-like globin gene cluster and DNase I HS2. The shaded and solid boxes are the sites of curved DNA determined by circular permutation assay and the DNA bend centers, respectively (32). The positions of the bend sites relative to the cap site of the ɛ-globin gene are: ɛB-17, –11 420 (SacI) to –11 166 (AflIII); ɛB-16, –10 558 (MunI) to –10 408 (EarI). The circles indicate the nucleosomal phases reported previously (32). (B) The mutant (Δb) was constructed by replacing the nucleosome regions containing the bend centers with fragments of identical length derived from pBR322 (positions 247–404 or positions 417–574 for ɛB-17 or ɛB-16, respectively) indicated as ‘Tc’ to remove bendability and nucleosome forming activity. The positions of the probes for Southern blot analysis correspond to the nucleosome positions. (C) Southern blot analysis of the in vitro reconstituted nucleosomes using plasmids, pHS2 and Δb. The plasmid DNA was digested with increasing amounts of micrococcal nuclease and the purified DNA was subjected to Southern blot analysis. The membrane was stripped and rehybridized with the probes shown as the horizontal arrows in (B). ‘Mono’ indicates the position of the mononucleosomes.