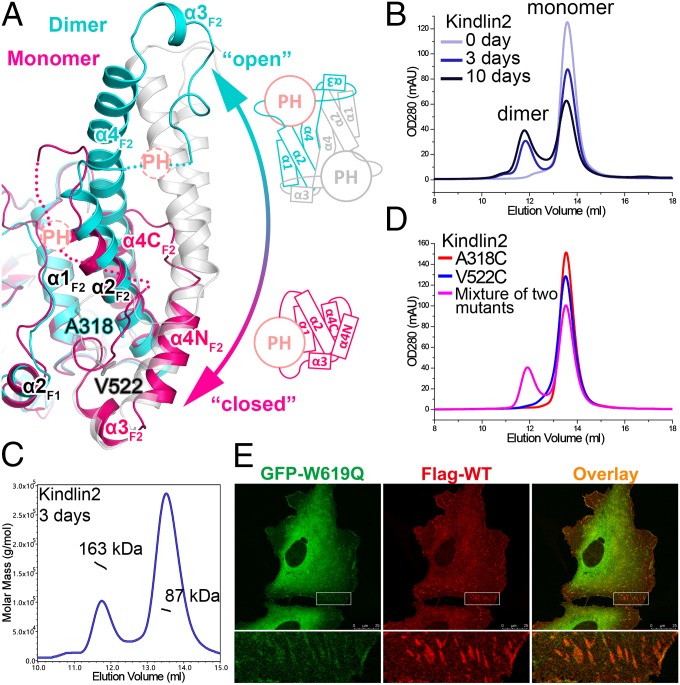
Fig. 4.
Structural basis of the monomer–dimer transition in kindlin2. (A) Structural comparison of the kindlin2 dimer and monomer. The insert PH domains and their connecting loops to the F2 lobes are indicated by dashed circles and lines, respectively. The open and closed conformations of the F2 lobe are also shown in the schematic views. The two residues, mutated to Cys, in D, are highlighted in the stick mode. (B) Analytical gel filtration analysis showing the dimer formation of kindlin2 in solution. The samples were prepared using the fresh-purified protein (0 d) or the same batch of protein placed at 4 °C for 3 d or 10 d. (C) The molecular weights of the dimer and monomer fractions were measured by using multiangle static light scattering. (D) Disulfide-bonding enhanced kindlin2 dimerization by structure-based mutation design. Before gel filtration analysis, the fresh-purified mutants and their mixture were kept at 4 °C overnight. (E) Cotransfection of the GFP-tagged W619Q mutant and the Flag-tagged wild-type protein in HT1080 cells. (Scale bar, 25 μm.)