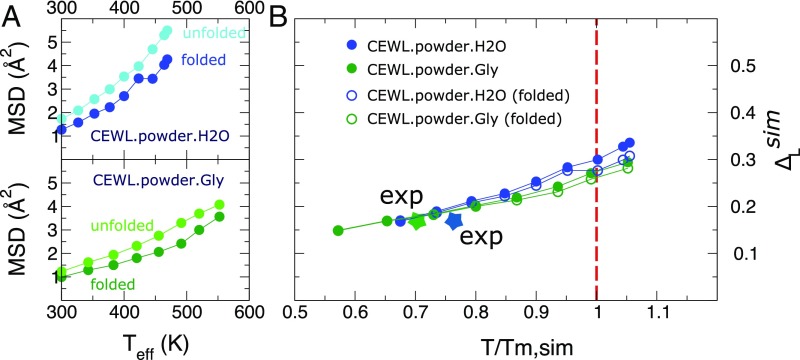
Fig. 4.
Scaling law of protein fast fluctuations by simulations. (A) MSD from MD trajectories started from folded and unfolded configurations of CEWL in the presence of water and glycerol. The data for the unfolded state are obtained as the average over four independent runs initiated from different configurations of the unfolded CEWL extracted from the REST2 enhanced sampling. (B) Lindemann parameter obtained by combining the folded and unfolded states MSD, , with the fraction of folded states. In the same graph we report the folded-state contributions. The colored stars indicate the experimental points for the two systems.