Figure 1. SGI-110 treatment induces hypomethylation and expression of NY-ESO-1 and MAGE-A3/6 in leukemic cell lines in vitro.
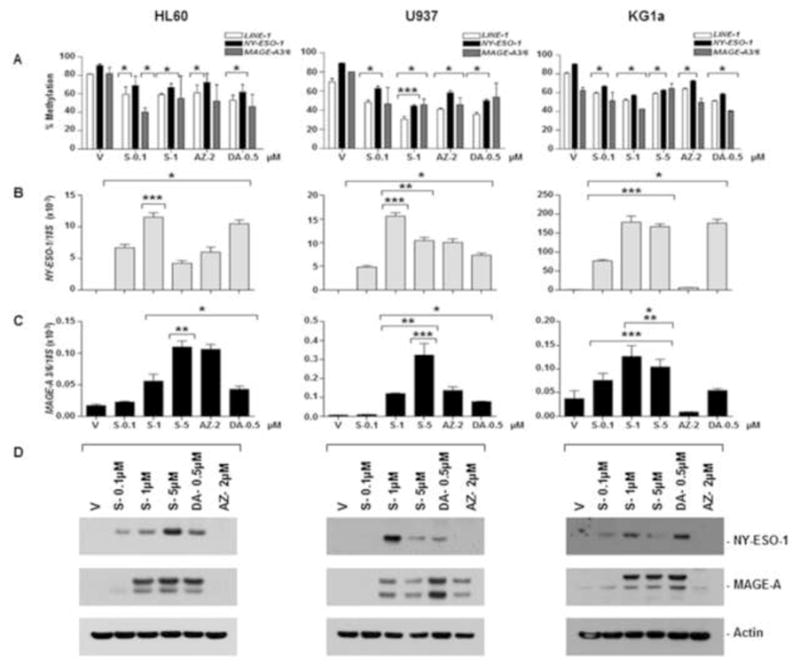
HL60, U937, or KG1a cells were treated with SGI-110, DAC and AZA as described. A) LINE-1 and NY-ESO-1 promoter methylation were determined by bisulfite pyrosequencing. MAGE-A3/6 methylation is expressed as a relative percentage of methylated product (MP/(UP+MP)*100) based upon densitometry from MSP (Representative of two experiments; primary data are shown in supplemental Figure 2) B) NY-ESO-1 and MAGE-A3/6 mRNA were quantified by qRT-PCR. C) NY-ESO-1 and MAGE-A protein expression was determined by western blot. All experiments were completed at least three times. * p<0.05 vs. vehicle, **p<0.05 vs. DAC, ***p<0.05 vs. AZA, S-SGI-110, V-Vehicle, DA-Decitabine, AZ-Azacytidine, MP-Density of methylated PCR product, UP- Density of unmethylated PCR product
