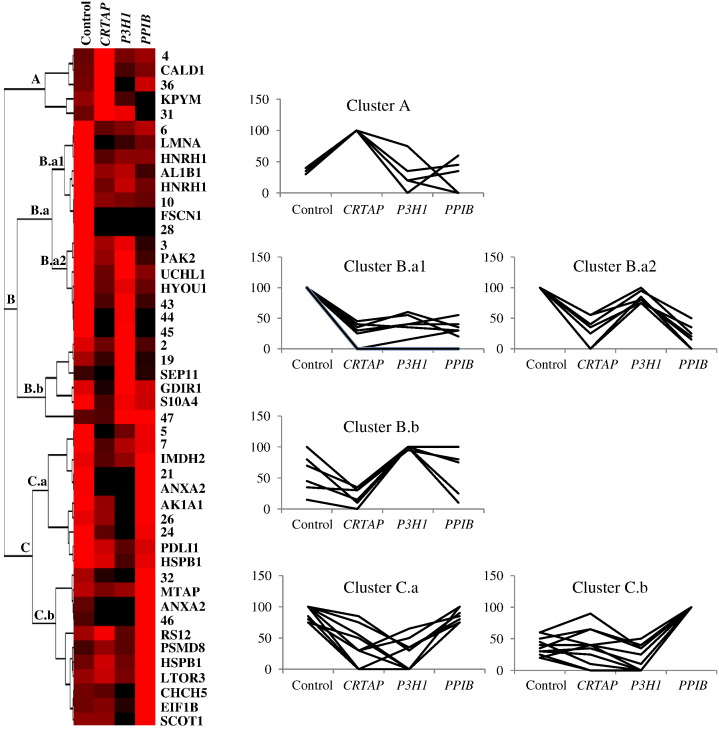
Fig. 2.
Unsupervised Hierarchical Clustering (HC) analysis. Normalized %V mean values of significant differences, as computed in spot maps from control, CRTAP, P3H1, and PPIB cells, are visualized by expression matrix, where each represented %V value is proportional to color intensity, from black = zero expression to red = maximum positive expression. Spot identity (protein name, as listed in Table 1, or number, for MS not identified spots) is annotated on the right of the expression matrix, while dendrogram of complete linkage clustering, based on centered correlation distance, is reported on the left. Main clusters are named from top to bottom and the corresponding expression profiles of their included proteins are shown on the right.