Figure 2.
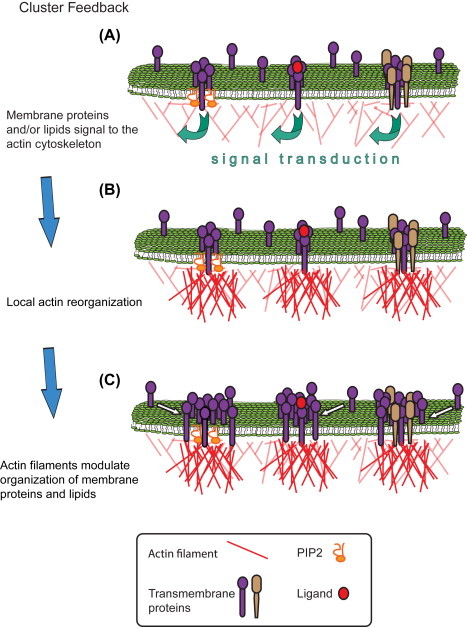
“Cluster Feedback” model of membrane organization. (A) Proteins cluster at the nanoscale in the plasma membrane. These clusters are protein–lipid (left, transmembrane protein and PIP2), mediated by ligand binding (middle), or protein–protein (right; depicted as a heterocluster; a homocluster oligomer is not shown). Of course, in clusters of any protein there exists the potential for local lipid clustering also. These protein–protein or protein–lipid clusters are collectively referred to hereafter as “membrane clusters.” In each case, signaling to the actin cytoskeleton initiated by proteins and/or lipids in membrane clusters elicits the local remodeling of actin (B), either through the recruitment and binding of actin filaments to the membrane lipids or proteins, the de novo nucleation of new filaments or branches thereof, or both. ABPs which mediate these interactions are not shown, but see Section 3.1 for more detail. The increase in actin density immediately adjacent to the membrane cluster then acts as a recruitment platform for other proteins (or lipids, not shown) diffusing in the membrane to join existing membrane clusters (C). This results in changes in the size, density, perimeter, and/or number of molecules within membrane clusters. Readers should note, the numbers of proteins depicted above are only for ease of communication—we do not hypothesize about the specific sizes (or numbers of molecules within) any of the membrane clusters here. Clusters may exist on the nano-, meso-, or micro scale; the Cluster Feedback model only predicts changes (here shown as increases) in cluster sizes from (A) to (C). See Sections 1.1 and 3.1 for more detail. (See color plate)
