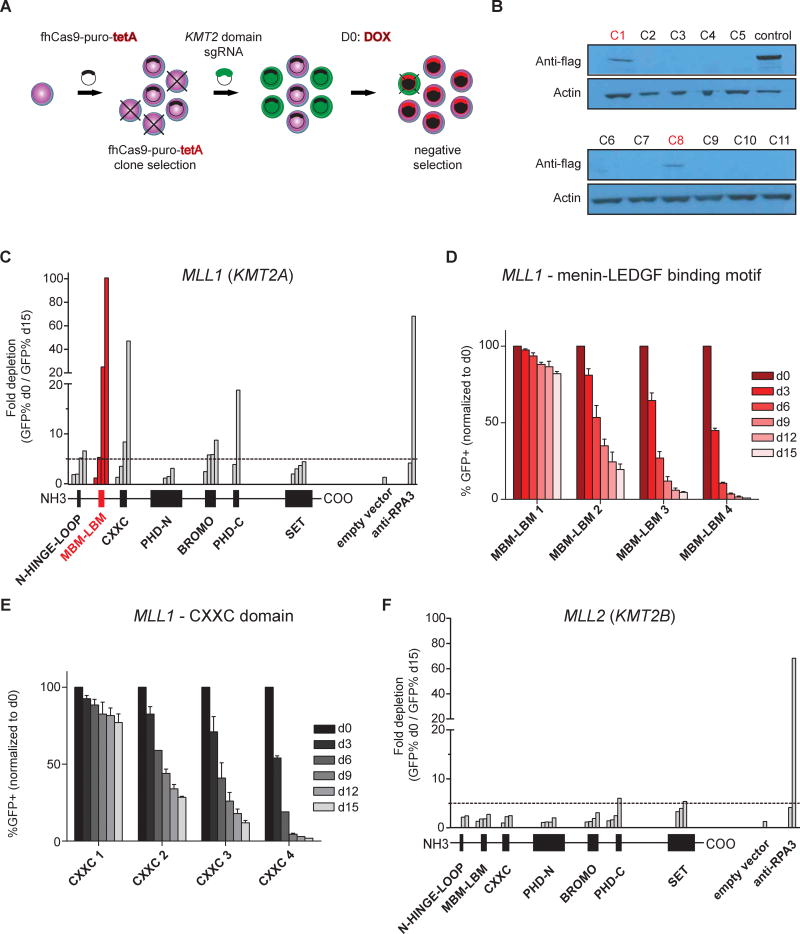
Figure 1. CRISPR-Cas9 mutagenesis of exons targeting MLL protein domains in NPM1mut AML cells.
(A) Experimental strategy for CRISPR-Cas9 negative selection screening: Engineering a clonal NPM1mut OCI-AML3 cell line that expresses a flag-tagged human codon-optimized Cas9 (fhCas9) vector containing a puro resistance gene (puro) and tetracycline-inducible transcriptional activator (tetA). GFP reporters of sgRNA constructs were used to track sgRNA negative selection after doxycycline induction of Cas9 (D0, day 0; DOX, doxycycline).
(B) Immunoblotting for flag-tagged hCas9 after doxycycline treatment in eleven OCI-AML3-Cas9 single cell clones. C1 and C8 clones were selected for two independent screens of MLL1 and MLL2.
(C and F) Summary of negative selection experiments with sgRNAs targeting exons encoding specific MLL1 and MLL2 protein domains. Negative selection is plotted as the fold depletion of GFP+ cells (d0 GFP% divided by d15 GFP%) during 18 days in culture. Each bar represents an independent sgRNA. The location of each sgRNA relative to the MLL1 or MLL2 protein is indicated along the x axis. The dashed line indicates a fivefold change. The data shown are the mean value of two independent replicates. Empty vector and anti-RPA3 sgRNA represent negative and positive controls.
(D and E) Negative selection competition assay that plots the percentage of GFP+ cells over time following transduction of OCI-AML3-Cas9 with the indicated sgRNAs. GFP+ percentage is normalized to the day 0 measurement following doxycycline induction of Cas9 (3 days after sgRNA transduction).
N-HINGE-LOOP, N-terminal hinge loop of the menin-LEDGF binding motif lacking many specific interactions; MBM-LBM, N-terminal fragment of MLL1 containing the menin and the LEDGF binding motif (as defined by Huang et al(35)); CXXC, CXXC-type zink finger domain; PHD-N, N-terminal plant homeodomains; BROMO, bromodomain; PHD-C , C-terminal plant homeodomain; SET, SET-domain. Data shown in panel C-F represent mean of biological duplicates.