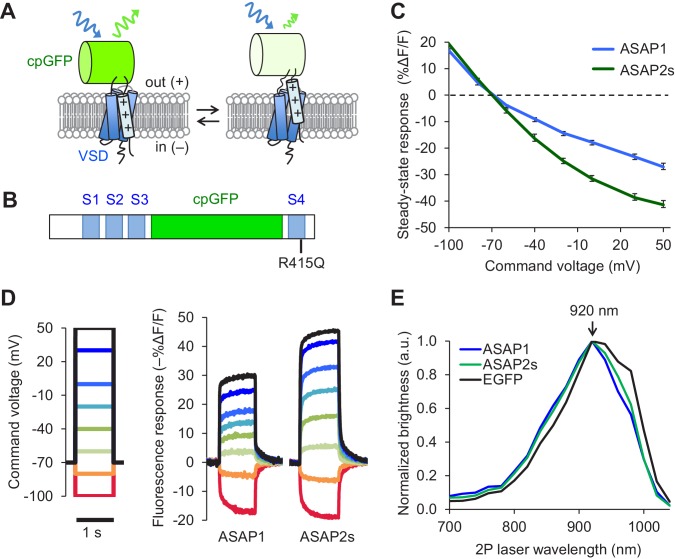
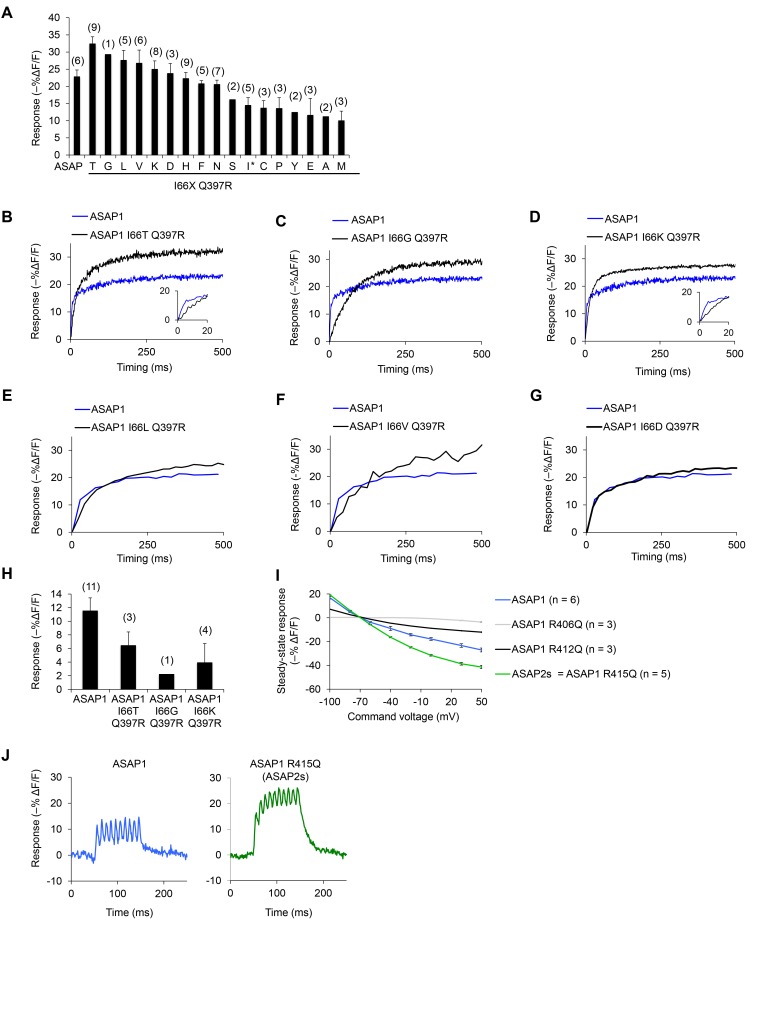
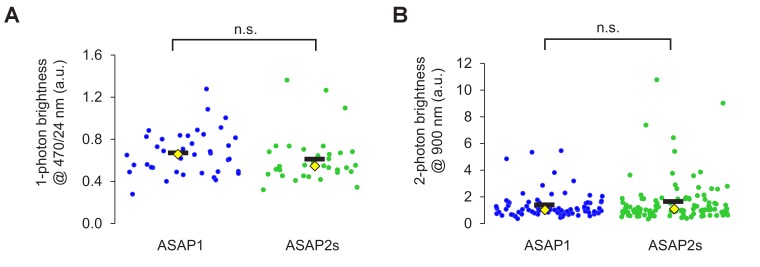
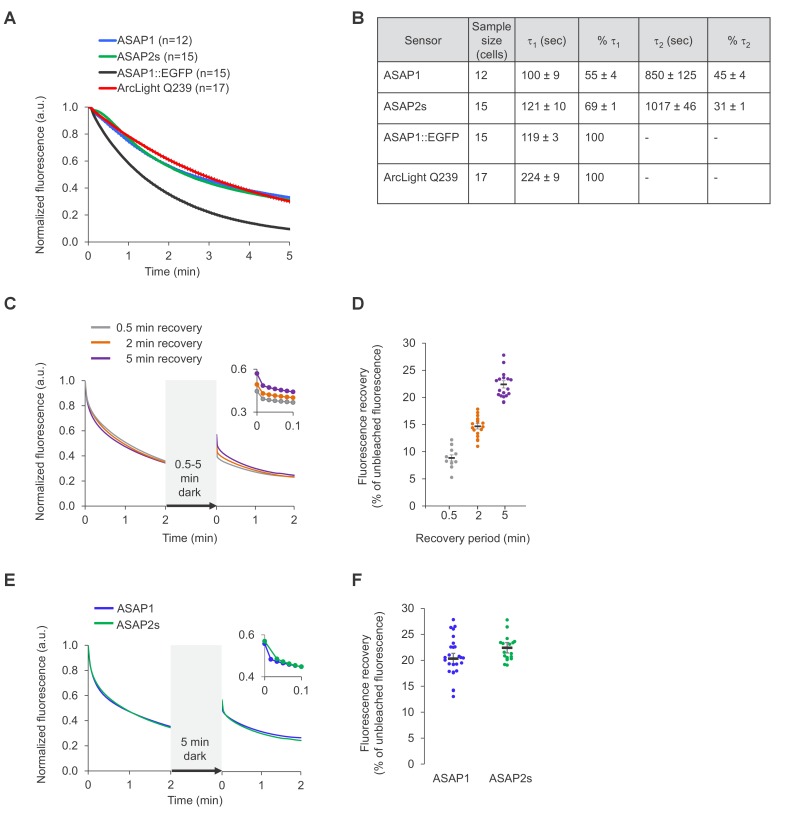
Figure 1. Design and in vitro characterization of ASAP2s.
(A) In ASAP-type sensors, voltage-induced movement of a positively charged transmembrane helix of a voltage-sensing domain (VSD) is thought to perturb the protonation state of a circularly permuted GFP (cpGFP), resulting in changes in fluorescence emission. (B) Schematic diagram of ASAP1, showing the VSD transmembrane domains (S1 to S4, blue), cpGFP, and the location of the new mutation in ASAP2s (R415Q). (C) Mean fluorescence responses to voltage steps (n = 6 HEK293A cells for ASAP1 and n = 5 for ASAP2s). Error bars are standard error of the mean (SEM). Responses are reported as the fluorescence change (ΔF) normalized by the initial fluorescence (F), expressed as a percentage of the initial fluorescence (% ΔF/F). (D) Representative fluorescence responses of ASAP1 and ASAP2s to voltage steps from −100 to 50 mV. Responses were measured at 5 ms intervals and were normalized to the fluorescence at the −70 mV holding potential. (E) Two-photon excitation spectra of ASAP1, ASAP2s, and EGFP. All proteins were expressed in HEK293-Kir2.1 cells with resting membrane potential of ~-77 mV. Brightness was evaluated every 20 nm from 700 to 1040 nm. Each spectrum was normalized to its peak brightness. Traces are the mean of >30 cells.