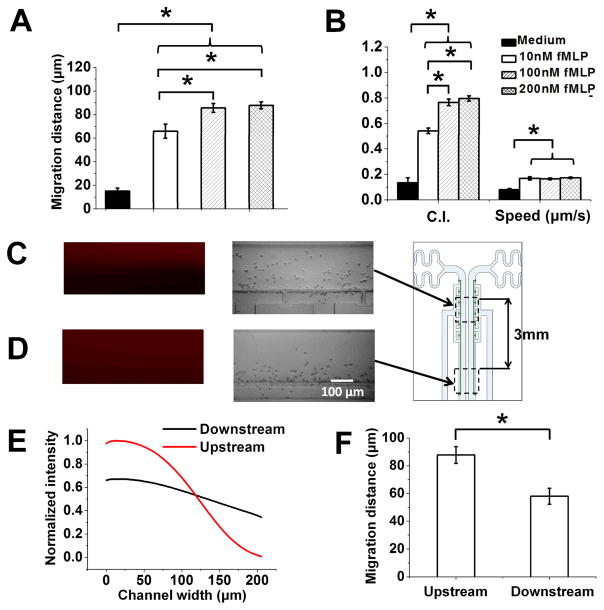
Fig. 6. Chemotaxis test of neutrophils to fMLP gradients of different doses and profiles using the Mkit.
(A) Cell migration distance of neutrophils to fMLP gradients of different doses; (B) Cell tracking analysis by calculating CI and cell speed to different fMLP gradient doses; (C–D) Rhodamine 6 G gradient images and the corresponding final cell distribution images at the upstream and downstream of the gradient channel; (E) Plot of gradient profiles at the upstream and downstream (normalized to the upstream gradient) of the gradient channel; (F) Cell migration distance analysis of neutrophil in upstream and downstream to a 100 nM fMLP gradient. The bars show the average value of the parameter of all cells for each condition and the error bars are the standard error of the mean (s.e.m.). The data shown are from a set of representative experiments (one experiment for each condition)